[English] 日本語
 Yorodumi
Yorodumi- EMDB-24239: High-resolution structure of photosystem II from the mesophilic c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24239 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | High-resolution structure of photosystem II from the mesophilic cyanobacterium, Synechocystis sp. PCC 6803 | |||||||||
 Map data Map data | Photosystem II from Synechocystis 6803 full dose unsharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Photosystem II / Cyanobacteria / Synechocystis 6803 / Mesophile / CyanoQ / PHOTOSYNTHESIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationplasma membrane-derived thylakoid photosystem II / photosystem II oxygen evolving complex / photosystem II assembly / oxygen evolving activity / photosystem II stabilization / photosystem II reaction center / photosystem II / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor / photosynthetic electron transport chain / : ...plasma membrane-derived thylakoid photosystem II / photosystem II oxygen evolving complex / photosystem II assembly / oxygen evolving activity / photosystem II stabilization / photosystem II reaction center / photosystem II / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor / photosynthetic electron transport chain / : / response to herbicide / photosystem II / extrinsic component of membrane / plasma membrane-derived thylakoid membrane / photosynthetic electron transport in photosystem II / chlorophyll binding / phosphate ion binding / photosynthesis, light reaction / photosynthesis / respiratory electron transport chain / manganese ion binding / electron transfer activity / protein stabilization / iron ion binding / heme binding / calcium ion binding / metal ion binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
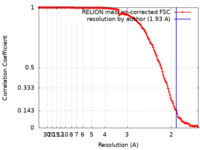
| Method | single particle reconstruction / cryo EM / Resolution: 1.93 Å | |||||||||
 Authors Authors | Gisriel CJ / Brudvig GW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: High-resolution cryo-electron microscopy structure of photosystem II from the mesophilic cyanobacterium, sp. PCC 6803. Authors: Christopher J Gisriel / Jimin Wang / Jinchan Liu / David A Flesher / Krystle M Reiss / Hao-Li Huang / Ke R Yang / William H Armstrong / M R Gunner / Victor S Batista / Richard J Debus / Gary W Brudvig /  Abstract: Photosystem II (PSII) enables global-scale, light-driven water oxidation. Genetic manipulation of PSII from the mesophilic cyanobacterium sp. PCC 6803 has provided insights into the mechanism of ...Photosystem II (PSII) enables global-scale, light-driven water oxidation. Genetic manipulation of PSII from the mesophilic cyanobacterium sp. PCC 6803 has provided insights into the mechanism of water oxidation; however, the lack of a high-resolution structure of oxygen-evolving PSII from this organism has limited the interpretation of biophysical data to models based on structures of thermophilic cyanobacterial PSII. Here, we report the cryo-electron microscopy structure of PSII from sp. PCC 6803 at 1.93-Å resolution. A number of differences are observed relative to thermophilic PSII structures, including the following: the extrinsic subunit PsbQ is maintained, the C terminus of the D1 subunit is flexible, some waters near the active site are partially occupied, and differences in the PsbV subunit block the Large (O1) water channel. These features strongly influence the structural picture of PSII, especially as it pertains to the mechanism of water oxidation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24239.map.gz emd_24239.map.gz | 29.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24239-v30.xml emd-24239-v30.xml emd-24239.xml emd-24239.xml | 39.1 KB 39.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24239_fsc.xml emd_24239_fsc.xml | 13.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_24239.png emd_24239.png | 52.8 KB | ||
| Filedesc metadata |  emd-24239.cif.gz emd-24239.cif.gz | 8.7 KB | ||
| Others |  emd_24239_additional_1.map.gz emd_24239_additional_1.map.gz | 169.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24239 http://ftp.pdbj.org/pub/emdb/structures/EMD-24239 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24239 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24239 | HTTPS FTP |
-Related structure data
| Related structure data |  7n8oMC  7rcvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24239.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24239.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Photosystem II from Synechocystis 6803 full dose unsharpened | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Photosystem II from Synechocystis 6803 full dose sharpened
| File | emd_24239_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Photosystem II from Synechocystis 6803 full dose sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Photosystem II
+Supramolecule #1: Photosystem II
+Macromolecule #1: Photosystem II protein D1 2
+Macromolecule #2: Photosystem II CP47 reaction center protein
+Macromolecule #3: Photosystem II CP43 reaction center protein
+Macromolecule #4: Photosystem II D2 protein
+Macromolecule #5: Cytochrome b559 subunit alpha
+Macromolecule #6: Cytochrome b559 subunit beta
+Macromolecule #7: Photosystem II reaction center protein H
+Macromolecule #8: Photosystem II reaction center protein I
+Macromolecule #9: Photosystem II reaction center protein J
+Macromolecule #10: Photosystem II reaction center protein K
+Macromolecule #11: Photosystem II reaction center protein L
+Macromolecule #12: Photosystem II reaction center protein M
+Macromolecule #13: Photosystem II manganese-stabilizing polypeptide
+Macromolecule #14: Sll1638 protein
+Macromolecule #15: Photosystem II protein Y
+Macromolecule #16: Photosystem II reaction center protein T
+Macromolecule #17: Photosystem II 12 kDa extrinsic protein
+Macromolecule #18: Cytochrome c-550
+Macromolecule #19: Photosystem II reaction center X protein
+Macromolecule #20: Photosystem II reaction center protein Ycf12
+Macromolecule #21: Photosystem II reaction center protein Z
+Macromolecule #22: CA-MN4-O5 CLUSTER
+Macromolecule #23: FE (II) ION
+Macromolecule #24: CHLORIDE ION
+Macromolecule #25: CHLOROPHYLL A
+Macromolecule #26: PHEOPHYTIN A
+Macromolecule #27: BETA-CAROTENE
+Macromolecule #28: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #29: 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,...
+Macromolecule #30: 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL
+Macromolecule #31: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #32: BICARBONATE ION
+Macromolecule #33: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #34: DIGALACTOSYL DIACYL GLYCEROL (DGDG)
+Macromolecule #35: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #36: CALCIUM ION
+Macromolecule #37: (3R)-beta,beta-caroten-3-ol
+Macromolecule #38: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)