[English] 日本語
 Yorodumi
Yorodumi- EMDB-24195: Complex structure of HIV superinfection Fab QA013.2 and BG505.SOS... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24195 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex structure of HIV superinfection Fab QA013.2 and BG505.SOSIP.664 | |||||||||
 Map data Map data | Complex structure of QA013.2 Fab and BG505.SOSIP.664 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 Envelope / superinfection antibody QA013.2 / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane fusion involved in viral entry into host cell / symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope ...membrane fusion involved in viral entry into host cell / symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
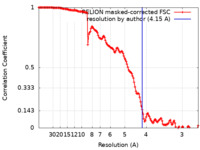
| Method | single particle reconstruction / cryo EM / Resolution: 4.15 Å | |||||||||
 Authors Authors | Mangala Prasad V / Shipley MM / Overbaugh JM / Lee KK | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Functional development of a V3/glycan-specific broadly neutralizing antibody isolated from a case of HIV superinfection. Authors: Mackenzie M Shipley / Vidya Mangala Prasad / Laura E Doepker / Adam Dingens / Duncan K Ralph / Elias Harkins / Amrit Dhar / Dana Arenz / Vrasha Chohan / Haidyn Weight / Kishor Mandaliya / ...Authors: Mackenzie M Shipley / Vidya Mangala Prasad / Laura E Doepker / Adam Dingens / Duncan K Ralph / Elias Harkins / Amrit Dhar / Dana Arenz / Vrasha Chohan / Haidyn Weight / Kishor Mandaliya / Jesse D Bloom / Frederick A Matsen / Kelly K Lee / Julie M Overbaugh /   Abstract: Stimulating broadly neutralizing antibodies (bnAbs) directly from germline remains a barrier for HIV vaccines. HIV superinfection elicits bnAbs more frequently than single infection, providing clues ...Stimulating broadly neutralizing antibodies (bnAbs) directly from germline remains a barrier for HIV vaccines. HIV superinfection elicits bnAbs more frequently than single infection, providing clues of how to elicit such responses. We used longitudinal antibody sequencing and structural studies to characterize bnAb development from a superinfection case. BnAb QA013.2 bound initial and superinfecting viral Env, despite its probable naive progenitor only recognizing the superinfecting strain, suggesting both viruses influenced this lineage. A 4.15 Å cryo-EM structure of QA013.2 bound to native-like trimer showed recognition of V3 signatures (N301/N332 and GDIR). QA013.2 relies less on CDRH3 and more on framework and CDRH1 for affinity and breadth compared to other V3/glycan-specific bnAbs. Antigenic profiling revealed that viral escape was achieved by changes in the structurally-defined epitope and by mutations in V1. These results highlight shared and novel properties of QA013.2 relative to other V3/glycan-specific bnAbs in the setting of sequential, diverse antigens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24195.map.gz emd_24195.map.gz | 228.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24195-v30.xml emd-24195-v30.xml emd-24195.xml emd-24195.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24195_fsc.xml emd_24195_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_24195.png emd_24195.png | 73.4 KB | ||
| Filedesc metadata |  emd-24195.cif.gz emd-24195.cif.gz | 7.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24195 http://ftp.pdbj.org/pub/emdb/structures/EMD-24195 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24195 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24195 | HTTPS FTP |
-Related structure data
| Related structure data |  7n65MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24195.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24195.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Complex structure of QA013.2 Fab and BG505.SOSIP.664 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of HIV-1 envelope protein with super-infection Fab QA013.2
| Entire | Name: Complex of HIV-1 envelope protein with super-infection Fab QA013.2 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of HIV-1 envelope protein with super-infection Fab QA013.2
| Supramolecule | Name: Complex of HIV-1 envelope protein with super-infection Fab QA013.2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: BG505 Human immunodeficiency virus 1 / Strain: BG505 |
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: BG505 Human immunodeficiency virus 1 / Strain: BG505 |
| Molecular weight | Theoretical: 56.846613 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAENLW VTVYYGVPVW KDAETTLFCA SDAKAYETEK HNVWATHACV PTDPNPQEI HLENVTEEFN MWKNNMVEQM HTDIISLWDQ SLKPCVKLTP LCVTLQCTNV TNNITDDMRG ELKNCSFNMT T ELRDKKQK ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAENLW VTVYYGVPVW KDAETTLFCA SDAKAYETEK HNVWATHACV PTDPNPQEI HLENVTEEFN MWKNNMVEQM HTDIISLWDQ SLKPCVKLTP LCVTLQCTNV TNNITDDMRG ELKNCSFNMT T ELRDKKQK VYSLFYRLDV VQINENQGNR SNNSNKEYRL INCNTSAITQ ACPKVSFEPI PIHYCAPAGF AILKCKDKKF NG TGPCPSV STVQCTHGIK PVVSTQLLLN GSLAEEEVMI RSENITNNAK NILVQFNTPV QINCTRPNNN TRKSIRIGPG QAF YATGDI IGDIRQAHCN VSKATWNETL GKVVKQLRKH FGNNTIIRFA NSSGGDLEVT THSFNCGGEF FYCNTSGLFN STWI SNTSV QGSNSTGSND SITLPCRIKQ IINMWQRIGQ AMYAPPIQGV IRCVSNITGL ILTRDGGSTN STTETFRPGG GDMRD NWRS ELYKYKVVKI EPLGVAPTRC KRRV UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: BG505 Human immunodeficiency virus 1 / Strain: BG505 |
| Molecular weight | Theoretical: 18.24583 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VGRRRRRRAV GIGAVFLGFL GAAGSTMGAA SMTLTVQARN LLSGIVQQQS NLLRAPEAQQ HLLKLTVWGI KQLQARVLAV ERYLRDQQL LGIWGCSGKL ICCTNVPWNS SWSNRNLSEI WDNMTWLQWD KEISNYTQII YGLLEESQNQ QEKNEQDLLA L D UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: Fab QA013.2 Heavy Chain, variable region
| Macromolecule | Name: Fab QA013.2 Heavy Chain, variable region / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.384085 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIRIAESGGG LVQPGESLRL ACEIIELGFR RAWTTWVRQA PGKGLEWVAD INEDGSEKKY GPSVTGRFTI SRDNGKNLVF LQMNSLRVE DTATYYCARE AYHLVYDDRI PRGNWFDPWG PGTLVTVSS |
-Macromolecule #4: Fab QA013.2 Light Chain, , variable region
| Macromolecule | Name: Fab QA013.2 Light Chain, , variable region / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.189263 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSV SGAPGQRVVI SCTGSRSNIG AGYDVHWYQQ SPGKVPRIII YGSNSRSSGV PARFSGSKSG TSASLAITGL QAEDEADYY CQSYDTTLTA SVFGGGTKV |
-Macromolecule #11: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 11 / Number of copies: 9 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #12: alpha-D-mannopyranose
| Macromolecule | Name: alpha-D-mannopyranose / type: ligand / ID: 12 / Number of copies: 3 / Formula: MAN |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-MAN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Name: Phosphate buffer saline / Details: 1X PBS at pH 7.4, made fresh. |
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 3ul sample volume, blotted for 3-4 seconds. |
| Details | Monodisperse sample in PBS |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | Preliminary grid screening was performed manually |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 6475 / Average exposure time: 10.0 sec. / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 110000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Rigid body fitting in Chimera, followed by real space refinement in Phenix using reference model restraints |
| Refinement | Protocol: RIGID BODY FIT |
| Output model |  PDB-7n65: |
 Movie
Movie Controller
Controller












 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)