+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24117 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Venezuelan equine encephalitis virus VLP | |||||||||
 Map data Map data | Sharpened cryo-electron microscopy map of Venezuelan equine encephalitis VLP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | viral envelope / encephalitic alphavirus / VLP / Structural Genomics / Center for Structural Genomics of Infectious Diseases / CSGID / VIRUS LIKE PARTICLE / VEEV | |||||||||
| Function / homology |  Function and homology information Function and homology informationT=4 icosahedral viral capsid / host cell cytoplasm / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / membrane Similarity search - Function | |||||||||
| Biological species |  Venezuelan equine encephalitis virus Venezuelan equine encephalitis virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Basore K / Nelson CA / Fremont DH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structure of Venezuelan equine encephalitis virus in complex with the LDLRAD3 receptor. Authors: Katherine Basore / Hongming Ma / Natasha M Kafai / Samantha Mackin / Arthur S Kim / Christopher A Nelson / Michael S Diamond / Daved H Fremont /  Abstract: LDLRAD3 is a recently defined attachment and entry receptor for Venezuelan equine encephalitis virus (VEEV), a New World alphavirus that causes severe neurological disease in humans. Here we present ...LDLRAD3 is a recently defined attachment and entry receptor for Venezuelan equine encephalitis virus (VEEV), a New World alphavirus that causes severe neurological disease in humans. Here we present near-atomic-resolution cryo-electron microscopy reconstructions of VEEV virus-like particles alone and in a complex with the ectodomains of LDLRAD3. Domain 1 of LDLRAD3 is a low-density lipoprotein receptor type-A module that binds to VEEV by wedging into a cleft created by two adjacent E2-E1 heterodimers in one trimeric spike, and engages domains A and B of E2 and the fusion loop in E1. Atomic modelling of this interface is supported by mutagenesis and anti-VEEV antibody binding competition assays. Notably, VEEV engages LDLRAD3 in a manner that is similar to the way that arthritogenic alphaviruses bind to the structurally unrelated MXRA8 receptor, but with a much smaller interface. These studies further elucidate the structural basis of alphavirus-receptor interactions, which could inform the development of therapies to mitigate infection and disease against multiple members of this family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24117.map.gz emd_24117.map.gz | 1.2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24117-v30.xml emd-24117-v30.xml emd-24117.xml emd-24117.xml | 24.8 KB 24.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24117_fsc.xml emd_24117_fsc.xml | 24.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_24117.png emd_24117.png | 278.9 KB | ||
| Filedesc metadata |  emd-24117.cif.gz emd-24117.cif.gz | 7.2 KB | ||
| Others |  emd_24117_additional_1.map.gz emd_24117_additional_1.map.gz emd_24117_half_map_1.map.gz emd_24117_half_map_1.map.gz emd_24117_half_map_2.map.gz emd_24117_half_map_2.map.gz | 1 GB 1 GB 1 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24117 http://ftp.pdbj.org/pub/emdb/structures/EMD-24117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24117 | HTTPS FTP |
-Validation report
| Summary document |  emd_24117_validation.pdf.gz emd_24117_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24117_full_validation.pdf.gz emd_24117_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_24117_validation.xml.gz emd_24117_validation.xml.gz | 32.3 KB | Display | |
| Data in CIF |  emd_24117_validation.cif.gz emd_24117_validation.cif.gz | 43.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24117 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24117 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24117 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24117 | HTTPS FTP |
-Related structure data
| Related structure data |  7n1iMC  7n1hC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24117.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24117.map.gz / Format: CCP4 / Size: 1.3 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened cryo-electron microscopy map of Venezuelan equine encephalitis VLP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
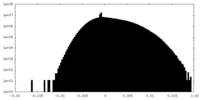
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unsharpened cryo-electron microscopy map of Venezuelan equine encephalitis...
| File | emd_24117_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened cryo-electron microscopy map of Venezuelan equine encephalitis VLP | ||||||||||||
| Projections & Slices |
| ||||||||||||
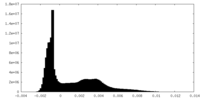
| Density Histograms |
-Half map: Unsharpened half map 2.
| File | emd_24117_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened half map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
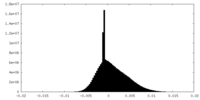
| Density Histograms |
-Half map: Unsharpened half map 1.
| File | emd_24117_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened half map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
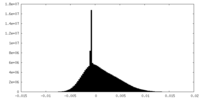
| Density Histograms |
- Sample components
Sample components
-Entire : Venezuelan equine encephalitis virus
| Entire | Name:  Venezuelan equine encephalitis virus Venezuelan equine encephalitis virus |
|---|---|
| Components |
|
-Supramolecule #1: Venezuelan equine encephalitis virus
| Supramolecule | Name: Venezuelan equine encephalitis virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Virus like particle produce in HEK293F cells by recombinant expression of structural proteins. NCBI-ID: 11036 / Sci species name: Venezuelan equine encephalitis virus / Sci species strain: TC-83 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  |
-Macromolecule #1: E1 envelope glycoprotein
| Macromolecule | Name: E1 envelope glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Venezuelan equine encephalitis virus Venezuelan equine encephalitis virus |
| Molecular weight | Theoretical: 47.952066 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: YEHATTMPSQ AGISYNTIVN RAGYAPLPIS ITPTKIKLIP TVNLEYVTCH YKTGMDSPAI KCCGSQECTP TYRPDEQCKV FTGVYPFMW GGAYCFCDTE NTQVSKAYVM KSDDCLADHA EAYKAHTASV QAFLNITVGE HSIVTTVYVN GETPVNFNGV K ITAGPLST ...String: YEHATTMPSQ AGISYNTIVN RAGYAPLPIS ITPTKIKLIP TVNLEYVTCH YKTGMDSPAI KCCGSQECTP TYRPDEQCKV FTGVYPFMW GGAYCFCDTE NTQVSKAYVM KSDDCLADHA EAYKAHTASV QAFLNITVGE HSIVTTVYVN GETPVNFNGV K ITAGPLST AWTPFDRKIV QYAGEIYNYD FPEYGAGQPG AFGDIQSRTV SSSDLYANTN LVLQRPKAGA IHVPYTQAPS GF EQWKKDK APSLKFTAPF GCEIYTNPIR AENCAVGSIP LAFDIPDALF TRVSETPTLS AAECTLNECV YSSDFGGIAT VKY SASKSG KCAVHVPSGT ATLKEAAVEL TEQGSATIHF STANIHPEFR LQICTSYVTC KGDCHPPKDH IVTHPQYHAQ TFTA AVSKT AWTWLTSLLG GSAVIIIIGL VLATIVAMYV LTNQKHN UniProtKB: Structural polyprotein |
-Macromolecule #2: E2 envelope glycoprotein
| Macromolecule | Name: E2 envelope glycoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Venezuelan equine encephalitis virus Venezuelan equine encephalitis virus |
| Molecular weight | Theoretical: 47.113746 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: STEELFNEYK LTRPYMARCI RCAVGSCHSP IAIEAVKSDG HDGYVRLQTS SQYGLDSSGN LKGRTMRYDM HGTIKEIPLH QVSLYTSRP CHIVDGHGYF LLARCPAGDS ITMEFKKDSV RHSCSVPYEV KFNPVGRELY THPPEHGVEQ ACQVYAHDAQ N RGAYVEMH ...String: STEELFNEYK LTRPYMARCI RCAVGSCHSP IAIEAVKSDG HDGYVRLQTS SQYGLDSSGN LKGRTMRYDM HGTIKEIPLH QVSLYTSRP CHIVDGHGYF LLARCPAGDS ITMEFKKDSV RHSCSVPYEV KFNPVGRELY THPPEHGVEQ ACQVYAHDAQ N RGAYVEMH LPGSEVDSSL VSLSGSSVTV TPPDGTSALV ECECGGTKIS ETINKTKQFS QCTKKEQCRA YRLQNDKWVY NS DKLPKAA GATLKGKLHV PFLLADGKCT VPLAPEPMIT FGFRSVSLKL HPKNPTYLIT RQLADEPHYT HELISEPAVR NFT VTEKGW EFVWGNHPPK RFWAQETAPG NPHGLPHEVI THYYHRYPMS TILGLSICAA IATVSVAAST WLFCRSRVAC LTPY RLTPN ARIPFCLAVL CCARTARA UniProtKB: Structural polyprotein |
-Macromolecule #3: Capsid
| Macromolecule | Name: Capsid / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Venezuelan equine encephalitis virus Venezuelan equine encephalitis virus |
| Molecular weight | Theoretical: 18.19584 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: KRQRMVMKLE SDKTFPIMLE GKINGYACVV GGKLFRPMHV EGKIDNDVLA ALKTKKASKY DLEYADVPQN MRADTFKYTH EKPQGYYSW HHGAVQYENG RFTVPKGVGA KGDSGRPILD NQGRVVAIVL GGVNEGSRTA LSVVMWNEKG VTVKYTPENC E QW UniProtKB: Structural polyprotein |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 8 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)