[English] 日本語
 Yorodumi
Yorodumi- EMDB-23915: Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23915 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / receptor-binding domain / antibody / IMMUNE SYSTEM-VIRAL PROTEIN complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.15 Å | |||||||||
 Authors Authors | Zhou T / Tsybovsky T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Authors: Lingshu Wang / Tongqing Zhou / Yi Zhang / Eun Sung Yang / Chaim A Schramm / Wei Shi / Amarendra Pegu / Olamide K Oloniniyi / Amy R Henry / Samuel Darko / Sandeep R Narpala / Christian ...Authors: Lingshu Wang / Tongqing Zhou / Yi Zhang / Eun Sung Yang / Chaim A Schramm / Wei Shi / Amarendra Pegu / Olamide K Oloniniyi / Amy R Henry / Samuel Darko / Sandeep R Narpala / Christian Hatcher / David R Martinez / Yaroslav Tsybovsky / Emily Phung / Olubukola M Abiona / Avan Antia / Evan M Cale / Lauren A Chang / Misook Choe / Kizzmekia S Corbett / Rachel L Davis / Anthony T DiPiazza / Ingelise J Gordon / Sabrina Helmold Hait / Tandile Hermanus / Prudence Kgagudi / Farida Laboune / Kwanyee Leung / Tracy Liu / Rosemarie D Mason / Alexandra F Nazzari / Laura Novik / Sarah O'Connell / Sijy O'Dell / Adam S Olia / Stephen D Schmidt / Tyler Stephens / Christopher D Stringham / Chloe Adrienna Talana / I-Ting Teng / Danielle A Wagner / Alicia T Widge / Baoshan Zhang / Mario Roederer / Julie E Ledgerwood / Tracy J Ruckwardt / Martin R Gaudinski / Penny L Moore / Nicole A Doria-Rose / Ralph S Baric / Barney S Graham / Adrian B McDermott / Daniel C Douek / Peter D Kwong / John R Mascola / Nancy J Sullivan / John Misasi /   Abstract: The emergence of highly transmissible SARS-CoV-2 variants of concern (VOCs) that are resistant to therapeutic antibodies highlights the need for continuing discovery of broadly reactive antibodies. ...The emergence of highly transmissible SARS-CoV-2 variants of concern (VOCs) that are resistant to therapeutic antibodies highlights the need for continuing discovery of broadly reactive antibodies. We identified four receptor binding domain-targeting antibodies from three early-outbreak convalescent donors with potent neutralizing activity against 23 variants, including the B.1.1.7, B.1.351, P.1, B.1.429, B.1.526, and B.1.617 VOCs. Two antibodies are ultrapotent, with subnanomolar neutralization titers [half-maximal inhibitory concentration (IC) 0.3 to 11.1 nanograms per milliliter; IC 1.5 to 34.5 nanograms per milliliter). We define the structural and functional determinants of binding for all four VOC-targeting antibodies and show that combinations of two antibodies decrease the in vitro generation of escape mutants, suggesting their potential in mitigating resistance development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23915.map.gz emd_23915.map.gz | 267.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23915-v30.xml emd-23915-v30.xml emd-23915.xml emd-23915.xml | 24.4 KB 24.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23915.png emd_23915.png | 114.8 KB | ||
| Masks |  emd_23915_msk_1.map emd_23915_msk_1.map | 536.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23915.cif.gz emd-23915.cif.gz | 7.8 KB | ||
| Others |  emd_23915_additional_1.map.gz emd_23915_additional_1.map.gz emd_23915_half_map_1.map.gz emd_23915_half_map_1.map.gz emd_23915_half_map_2.map.gz emd_23915_half_map_2.map.gz | 506.4 MB 498.4 MB 498.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23915 http://ftp.pdbj.org/pub/emdb/structures/EMD-23915 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23915 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23915 | HTTPS FTP |
-Related structure data
| Related structure data |  7mm0MC  7lrsC  7lrtC  7mlzC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23915.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23915.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.873 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
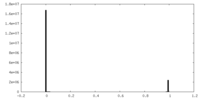
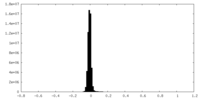
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23915_msk_1.map emd_23915_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
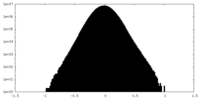
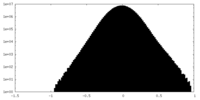
| Density Histograms |
-Additional map: #1
| File | emd_23915_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_23915_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_23915_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1
| Entire | Name: SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1
| Supramolecule | Name: SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Complex prepared by mixing SARS-CoV-2 spike protein and Fab fragment of antibody at a molar ratio of 1:3.6. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 539.951 KDa |
-Macromolecule #1: B1-182.1 Fab heavy chain
| Macromolecule | Name: B1-182.1 Fab heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.285312 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QMQLVQSGPE VKKPGTSVKV SCKASGFTFT SSAVQWVRQA RGQRLEWIGW IVVGSGNTNY AQKFQERVTI TRDMSTSTAY MELSSLRSE DTAVYYCAAP YCSGGSCFDG FDIWGQGTMV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: QMQLVQSGPE VKKPGTSVKV SCKASGFTFT SSAVQWVRQA RGQRLEWIGW IVVGSGNTNY AQKFQERVTI TRDMSTSTAY MELSSLRSE DTAVYYCAAP YCSGGSCFDG FDIWGQGTMV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSCDK |
-Macromolecule #2: B1-182.1 Fab light chain
| Macromolecule | Name: B1-182.1 Fab light chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Cell: B cells Homo sapiens (human) / Cell: B cells |
| Molecular weight | Theoretical: 23.536008 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRASQSVS SSYLAWYQQK PGQAPRLLIY GASSRATGFP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQYGNSPWTF GQGTKVEIRR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: EIVLTQSPGT LSLSPGERAT LSCRASQSVS SSYLAWYQQK PGQAPRLLIY GASSRATGFP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQYGNSPWTF GQGTKVEIRR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #3: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 132.334469 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHAIHVSGT NGTKRFDNPV LPFNDGVYFA STEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLG VYYHKNNKSW MESEFRVYSS ANNCTFEYVS Q PFLMDLEG ...String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHAIHVSGT NGTKRFDNPV LPFNDGVYFA STEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLG VYYHKNNKSW MESEFRVYSS ANNCTFEYVS Q PFLMDLEG KQGNFKNLRE FVFKNIDGYF KIYSKHTPIN LVRDLPQGFS ALEPLVDLPI GINITRFQTL LALHRSYLTP GD SSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPTESIVRFP NIT NLCPFG EVFNATRFAS VYAWNRKRIS NCVADYSVLY NSASFSTFKC YGVSPTKLND LCFTNVYADS FVIRGDEVRQ IAPG QTGKI ADYNYKLPDD FTGCVIAWNS NNLDSKVGGN YNYLYRLFRK SNLKPFERDI STEIYQAGST PCNGVEGFNC YFPLQ SYGF QPTNGVGYQP YRVVVLSFEL LHAPATVCGP KKSTNLVKNK CVNFNFNGLT GTGVLTESNK KFLPFQQFGR DIADTT DAV RDPQTLEILD ITPCSFGGVS VITPGTNTSN QVAVLYQDVN CTEVPVAIHA DQLTPTWRVY STGSNVFQTR AGCLIGA EH VNNSYECDIP IGAGICASYQ TQTNSPGSAS SVASQSIIAY TMSLGAENSV AYSNNSIAIP TNFTISVTTE ILPVSMTK T SVDCTMYICG DSTECSNLLL QYGSFCTQLN RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KDFGGFNFSQ ILPDPSKPS KRSPIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFNGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGPA LQIPFPMQM AYRFNGIGVT QNVLYENQKL IANQFNSAIG KIQDSLSSTP SALGKLQDVV NQNAQALNTL VKQLSSNFGA I SSVLNDIL SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FCGKGYHLMS FP QSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVSGNCDVVI GIV NNTVYD PLQPELDSFK EELDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQELGKYEQ UniProtKB: Spike glycoprotein |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 27 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 100 mM HEPES, 150 mM NaCl |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 4 seconds before plugging.. |
| Details | Complex at 0.5 mg/mL concentration in the presence of 0.005% DDM |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: OTHER / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Iterative refinement and manual model building/adjustment with Phenix and Coot. |
| Refinement | Space: REAL / Protocol: OTHER |
| Output model |  PDB-7mm0: |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)