[English] 日本語
 Yorodumi
Yorodumi- EMDB-23890: Straight tau filament extracted from PrP-CAA Patient brain tissue... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23890 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Straight tau filament extracted from PrP-CAA Patient brain tissue | tau filament | SF Tau | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology | Activation of AMPK downstream of NMDARs / PKR-mediated signaling / Isoform Tau-F of Microtubule-associated protein tau Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
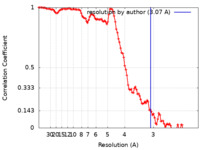
| Method | helical reconstruction / cryo EM / Resolution: 3.07 Å | |||||||||
 Authors Authors | Hoq MR | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Acta Neuropathol / Year: 2021 Journal: Acta Neuropathol / Year: 2021Title: Structure of Tau filaments in Prion protein amyloidoses. Authors: Grace I Hallinan / Md Rejaul Hoq / Manali Ghosh / Frank S Vago / Anllely Fernandez / Holly J Garringer / Ruben Vidal / Wen Jiang / Bernardino Ghetti /  Abstract: In human neurodegenerative diseases associated with the intracellular aggregation of Tau protein, the ordered cores of Tau filaments adopt distinct folds. Here, we analyze Tau filaments isolated from ...In human neurodegenerative diseases associated with the intracellular aggregation of Tau protein, the ordered cores of Tau filaments adopt distinct folds. Here, we analyze Tau filaments isolated from the brain of individuals affected by Prion-Protein cerebral amyloid angiopathy (PrP-CAA) with a nonsense mutation in the PRNP gene that leads to early termination of translation of PrP (Q160Ter or Q160X), and Gerstmann-Sträussler-Scheinker (GSS) disease, with a missense mutation in the PRNP gene that leads to an amino acid substitution at residue 198 (F198S) of PrP. The clinical and neuropathologic phenotypes associated with these two mutations in PRNP are different; however, the neuropathologic analyses of these two genetic variants have consistently shown the presence of numerous neurofibrillary tangles (NFTs) made of filamentous Tau aggregates in neurons. We report that Tau filaments in PrP-CAA (Q160X) and GSS (F198S) are composed of 3-repeat and 4-repeat Tau isoforms, having a striking similarity to NFTs in Alzheimer disease (AD). In PrP-CAA (Q160X), Tau filaments are made of both paired helical filaments (PHFs) and straight filaments (SFs), while in GSS (F198S), only PHFs were found. Mass spectrometry analyses of Tau filaments extracted from PrP-CAA (Q160X) and GSS (F198S) brains show the presence of post-translational modifications that are comparable to those seen in Tau aggregates from AD. Cryo-EM analysis reveals that the atomic models of the Tau filaments obtained from PrP-CAA (Q160X) and GSS (F198S) are identical to those of the Tau filaments from AD, and are therefore distinct from those of Pick disease, chronic traumatic encephalopathy, and corticobasal degeneration. Our data support the hypothesis that in the presence of extracellular amyloid deposits and regardless of the primary amino acid sequence of the amyloid protein, similar molecular mechanisms are at play in the formation of identical Tau filaments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23890.map.gz emd_23890.map.gz | 26.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23890-v30.xml emd-23890-v30.xml emd-23890.xml emd-23890.xml | 9.4 KB 9.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23890_fsc.xml emd_23890_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_23890.png emd_23890.png | 139 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23890 http://ftp.pdbj.org/pub/emdb/structures/EMD-23890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23890 | HTTPS FTP |
-Related structure data
| Related structure data |  7mkgMC  7mkfC  7mkhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23890.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23890.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.078 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Paired helical filament (PHF) from PrP-CAA
| Entire | Name: Paired helical filament (PHF) from PrP-CAA |
|---|---|
| Components |
|
-Supramolecule #1: Paired helical filament (PHF) from PrP-CAA
| Supramolecule | Name: Paired helical filament (PHF) from PrP-CAA / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: PHF tau in PrP-CAA, caused by the Q160X truncating mutation in PRNP |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Experimental: 55 kDa/nm |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Sugar embedding | Material: affinity tag |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: GATAN CRYOPLUNGE 3 |
| Details | Filament extracted from frontal cortex of PrP-CAA patient brain |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 2004 / Average electron dose: 1.067 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Details | Each model was refined using Rosetta |
|---|---|
| Refinement | Space: RECIPROCAL / Overall B value: 24 / Target criteria: Fourier shell correlation |
| Output model |  PDB-7mkg: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)