+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-23786 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 6.0 | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | nsp15 / sars-cov-2 / nendoU / coronavirus / covid-19 / VIRAL PROTEIN / HYDROLASE | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報protein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / mRNA guanylyltransferase activity / 5'-3' RNA helicase activity / 付加脱離酵素(リアーゼ); P-Oリアーゼ類; - / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / TRAF3-dependent IRF activation pathway / ISG15-specific peptidase activity ...protein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / mRNA guanylyltransferase activity / 5'-3' RNA helicase activity / 付加脱離酵素(リアーゼ); P-Oリアーゼ類; - / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / TRAF3-dependent IRF activation pathway / ISG15-specific peptidase activity / Transcription of SARS-CoV-2 sgRNAs / Translation of Replicase and Assembly of the Replication Transcription Complex / snRNP Assembly / Replication of the SARS-CoV-2 genome / 加水分解酵素; エステル加水分解酵素; 5'-リン酸モノエステル産生エキソリボヌクレアーゼ / host cell endoplasmic reticulum-Golgi intermediate compartment / double membrane vesicle viral factory outer membrane / SARS coronavirus main proteinase / 5'-3' DNA helicase activity / 3'-5'-RNA exonuclease activity / host cell endosome / symbiont-mediated degradation of host mRNA / mRNA guanylyltransferase / symbiont-mediated suppression of host ISG15-protein conjugation / G-quadruplex RNA binding / symbiont-mediated suppression of host toll-like receptor signaling pathway / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / omega peptidase activity / mRNA (guanine-N7)-methyltransferase / SARS-CoV-2 modulates host translation machinery / methyltransferase cap1 / host cell Golgi apparatus / symbiont-mediated suppression of host NF-kappaB cascade / symbiont-mediated perturbation of host ubiquitin-like protein modification / DNA helicase / methyltransferase cap1 activity / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / 加水分解酵素; プロテアーゼ; ペプチド結合加水分解酵素; システインプロテアーゼ / single-stranded RNA binding / host cell perinuclear region of cytoplasm / regulation of autophagy / viral protein processing / lyase activity / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / symbiont-mediated suppression of host gene expression / copper ion binding / viral translational frameshifting / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / lipid binding / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell nucleus / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane 類似検索 - 分子機能 | |||||||||
| 生物種 |  | |||||||||
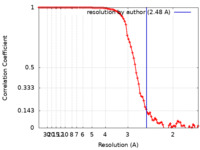
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.48 Å | |||||||||
 データ登録者 データ登録者 | Godoy AS / Song Y | |||||||||
| 資金援助 |  ブラジル, 1件 ブラジル, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nucleic Acids Res / 年: 2023 ジャーナル: Nucleic Acids Res / 年: 2023タイトル: Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease. 著者: Andre Schutzer Godoy / Aline Minalli Nakamura / Alice Douangamath / Yun Song / Gabriela Dias Noske / Victor Oliveira Gawriljuk / Rafaela Sachetto Fernandes / Humberto D Muniz Pereira / ...著者: Andre Schutzer Godoy / Aline Minalli Nakamura / Alice Douangamath / Yun Song / Gabriela Dias Noske / Victor Oliveira Gawriljuk / Rafaela Sachetto Fernandes / Humberto D Muniz Pereira / Ketllyn Irene Zagato Oliveira / Daren Fearon / Alexandre Dias / Tobias Krojer / Michael Fairhead / Alisa Powell / Louise Dunnet / Jose Brandao-Neto / Rachael Skyner / Rod Chalk / Dávid Bajusz / Miklós Bege / Anikó Borbás / György Miklós Keserű / Frank von Delft / Glaucius Oliva /      要旨: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19). The NSP15 endoribonuclease enzyme, known as NendoU, is highly conserved and ...Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19). The NSP15 endoribonuclease enzyme, known as NendoU, is highly conserved and plays a critical role in the ability of the virus to evade the immune system. NendoU is a promising target for the development of new antiviral drugs. However, the complexity of the enzyme's structure and kinetics, along with the broad range of recognition sequences and lack of structural complexes, hampers the development of inhibitors. Here, we performed enzymatic characterization of NendoU in its monomeric and hexameric form, showing that hexamers are allosteric enzymes with a positive cooperative index, and with no influence of manganese on enzymatic activity. Through combining cryo-electron microscopy at different pHs, X-ray crystallography and biochemical and structural analysis, we showed that NendoU can shift between open and closed forms, which probably correspond to active and inactive states, respectively. We also explored the possibility of NendoU assembling into larger supramolecular structures and proposed a mechanism for allosteric regulation. In addition, we conducted a large fragment screening campaign against NendoU and identified several new allosteric sites that could be targeted for the development of new inhibitors. Overall, our findings provide insights into the complex structure and function of NendoU and offer new opportunities for the development of inhibitors. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_23786.map.gz emd_23786.map.gz | 58.5 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-23786-v30.xml emd-23786-v30.xml emd-23786.xml emd-23786.xml | 18.3 KB 18.3 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_23786_fsc.xml emd_23786_fsc.xml | 8.9 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_23786.png emd_23786.png | 32.2 KB | ||
| マスクデータ |  emd_23786_msk_1.map emd_23786_msk_1.map | 64 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-23786.cif.gz emd-23786.cif.gz | 5.8 KB | ||
| その他 |  emd_23786_additional_1.map.gz emd_23786_additional_1.map.gz emd_23786_half_map_1.map.gz emd_23786_half_map_1.map.gz emd_23786_half_map_2.map.gz emd_23786_half_map_2.map.gz | 31.8 MB 59.3 MB 59.3 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23786 http://ftp.pdbj.org/pub/emdb/structures/EMD-23786 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23786 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23786 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_23786_validation.pdf.gz emd_23786_validation.pdf.gz | 934.5 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_23786_full_validation.pdf.gz emd_23786_full_validation.pdf.gz | 934 KB | 表示 | |
| XML形式データ |  emd_23786_validation.xml.gz emd_23786_validation.xml.gz | 16.7 KB | 表示 | |
| CIF形式データ |  emd_23786_validation.cif.gz emd_23786_validation.cif.gz | 21.3 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23786 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23786 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23786 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23786 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7me0MC  5s6xC  5s6yC  5s6zC  5s70C  5s71C  5s72C  5sa4C  5sa5C  5sa6C  5sa7C  5sa8C  5sa9C  5saaC  5sabC  5sacC  5sadC  5saeC  5safC  5sagC  5sahC  5saiC  5sbfC  7kegC  7kehC  7kf4C  7n7rC  7n7uC  7n7wC  7n7yC  7n83C  7rb0C  7rb2C M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | |
| 電子顕微鏡画像生データ |  EMPIAR-10794 (タイトル: Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 6.0 EMPIAR-10794 (タイトル: Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 6.0Data size: 359.3 Data #1: Aligned micrographs of SARS-CoV-2 NSP15 NendoU at pH 6.0 [micrographs - single frame]) |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_23786.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_23786.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.831 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-マスク #1
| ファイル |  emd_23786_msk_1.map emd_23786_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-追加マップ: #1
| ファイル | emd_23786_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_23786_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #2
| ファイル | emd_23786_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Structure of SARS-CoV-2 NSP15
| 全体 | 名称: Structure of SARS-CoV-2 NSP15 |
|---|---|
| 要素 |
|
-超分子 #1: Structure of SARS-CoV-2 NSP15
| 超分子 | 名称: Structure of SARS-CoV-2 NSP15 / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  |
-分子 #1: Uridylate-specific endoribonuclease
| 分子 | 名称: Uridylate-specific endoribonuclease / タイプ: protein_or_peptide / ID: 1 / コピー数: 6 / 光学異性体: LEVO EC番号: 加水分解酵素; エステル加水分解酵素 |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 42.117844 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MKHHHHHHPM SDYDIPTTEN LYFQGAMSLE NVAFNVVNKG HFDGQQGEVP VSIINNTVYT KVDGVDVELF ENKTTLPVNV AFELWAKRN IKPVPEVKIL NNLGVDIAAN TVIWDYKRDA PAHISTIGVC SMTDIAKKPT ETICAPLTVF FDGRVDGQVD L FRNARNGV ...文字列: MKHHHHHHPM SDYDIPTTEN LYFQGAMSLE NVAFNVVNKG HFDGQQGEVP VSIINNTVYT KVDGVDVELF ENKTTLPVNV AFELWAKRN IKPVPEVKIL NNLGVDIAAN TVIWDYKRDA PAHISTIGVC SMTDIAKKPT ETICAPLTVF FDGRVDGQVD L FRNARNGV LITEGSVKGL QPSVGPKQAS LNGVTLIGEA VKTQFNYYKK VDGVVQQLPE TYFTQSRNLQ EFKPRSQMEI DF LELAMDE FIERYKLEGY AFEHIVYGDF SHSQLGGLHL LIGLAKRFKE SPFELEDFIP MDSTVKNYFI TDAQTGSSKC VCS VIDLLL DDFVEIIKSQ DLSVVSKVVK VTIDYTEISF MLWCKDGHVE TFYPKLQ UniProtKB: Replicase polyprotein 1ab |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.5 mg/mL |
|---|---|
| 緩衝液 | pH: 6 |
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: COPPER / メッシュ: 300 / 前処理 - タイプ: GLOW DISCHARGE |
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 平均電子線量: 41.70522228 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー

























 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)