+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23784 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | LolCDE nucleotide-free | |||||||||
 Map data Map data | Cryo-EM map of LolCDE in the nucleotide-bound conformation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATP binding cassette transporter / ABC transporter / inner membrane / transport protein / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein localization to outer membrane / lipoprotein localization to outer membrane / plasma membrane protein complex / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / lipoprotein transport / transmembrane transporter activity / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Sharma S / Liao M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins. Authors: Stuti Sharma / Ruoyu Zhou / Li Wan / Shan Feng / KangKang Song / Chen Xu / Yanyan Li / Maofu Liao /   Abstract: Lipoproteins are important for bacterial growth and antibiotic resistance. These proteins use lipid acyl chains attached to the N-terminal cysteine residue to anchor on the outer surface of ...Lipoproteins are important for bacterial growth and antibiotic resistance. These proteins use lipid acyl chains attached to the N-terminal cysteine residue to anchor on the outer surface of cytoplasmic membrane. In Gram-negative bacteria, many lipoproteins are transported to the outer membrane (OM), a process dependent on the ATP-binding cassette (ABC) transporter LolCDE which extracts the OM-targeted lipoproteins from the cytoplasmic membrane. Lipid-anchored proteins pose a unique challenge for transport machinery as they have both hydrophobic lipid moieties and soluble protein component, and the underlying mechanism is poorly understood. Here we determined the cryo-EM structures of nanodisc-embedded LolCDE in the nucleotide-free and nucleotide-bound states at 3.8-Å and 3.5-Å resolution, respectively. The structural analyses, together with biochemical and mutagenesis studies, uncover how LolCDE recognizes its substrate by interacting with the lipid and N-terminal peptide moieties of the lipoprotein, and identify the amide-linked acyl chain as the key element for LolCDE interaction. Upon nucleotide binding, the transmembrane helices and the periplasmic domains of LolCDE undergo large-scale, asymmetric movements, resulting in extrusion of the captured lipoprotein. Comparison of LolCDE and MacB reveals the conserved mechanism of type VII ABC transporters and emphasizes the unique properties of LolCDE as a molecule extruder of triacylated lipoproteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23784.map.gz emd_23784.map.gz | 25.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23784-v30.xml emd-23784-v30.xml emd-23784.xml emd-23784.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23784.png emd_23784.png | 43.7 KB | ||
| Filedesc metadata |  emd-23784.cif.gz emd-23784.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23784 http://ftp.pdbj.org/pub/emdb/structures/EMD-23784 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23784 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23784 | HTTPS FTP |
-Validation report
| Summary document |  emd_23784_validation.pdf.gz emd_23784_validation.pdf.gz | 562 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23784_full_validation.pdf.gz emd_23784_full_validation.pdf.gz | 561.6 KB | Display | |
| Data in XML |  emd_23784_validation.xml.gz emd_23784_validation.xml.gz | 5.6 KB | Display | |
| Data in CIF |  emd_23784_validation.cif.gz emd_23784_validation.cif.gz | 6.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23784 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23784 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23784 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23784 | HTTPS FTP |
-Related structure data
| Related structure data |  7mdyMC  7mdxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23784.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23784.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of LolCDE in the nucleotide-bound conformation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.087 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of LolC, LolE and LolD in a 1:1:2 stoichiometry
| Entire | Name: Complex of LolC, LolE and LolD in a 1:1:2 stoichiometry |
|---|---|
| Components |
|
-Supramolecule #1: Complex of LolC, LolE and LolD in a 1:1:2 stoichiometry
| Supramolecule | Name: Complex of LolC, LolE and LolD in a 1:1:2 stoichiometry type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 130 kDa/nm |
-Macromolecule #1: Lipoprotein transporter subunit LolE
| Macromolecule | Name: Lipoprotein transporter subunit LolE / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.385977 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAMPLSLLIG LRFSRGRRRG GMVSLISVIS TIGIALGVAV LIVGLSAMNG FERELNNRIL AVVPHGEIEA VDQPWTNWQE ALDHVQKVP GIAAAAPYIN FTGLVESGAN LRAIQVKGVN PQQEQRLSAL PSFVQGDAWR NFKAGEQQII IGKGVADALK V KQGDWVSI ...String: MAMPLSLLIG LRFSRGRRRG GMVSLISVIS TIGIALGVAV LIVGLSAMNG FERELNNRIL AVVPHGEIEA VDQPWTNWQE ALDHVQKVP GIAAAAPYIN FTGLVESGAN LRAIQVKGVN PQQEQRLSAL PSFVQGDAWR NFKAGEQQII IGKGVADALK V KQGDWVSI MIPNSNPEHK LMQPKRVRLH VAGILQLSGQ LDHSFAMIPL ADAQQYLDMG SSVSGIALKM TDVFNANKLV RD AGEVTNS YVYIKSWIGT YGYMYRDIQM IRAIMYLAMV LVIGVACFNI VSTLVMAVKD KSGDIAVLRT LGAKDGLIRA IFV WYGLLA GLFGSLCGVI IGVVVSLQLT PIIEWIEKLI GHQFLSSDIY FIDFLPSELH WLDVFYVLVT ALLLSLLASW YPAR RASNI DPARVLSGQ UniProtKB: Lipoprotein transporter subunit LolE |
-Macromolecule #2: Lipo-releasing system transmembrane protein lolC
| Macromolecule | Name: Lipo-releasing system transmembrane protein lolC / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.295516 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYQPVALFIG LRYMRGRAAD RFGRFVSWLS TIGITLGVMA LVTVLSVMNG FERELQNNIL GLMPQAILSS EHGSLNPQQL PETAVKLDG VNRVAPITTG DVVLQSARSV AVGVMLGIDP AQKDPLTPYL VNVKQTDLEP GKYNVILGEQ LASQLGVNRG D QIRVMVPS ...String: MYQPVALFIG LRYMRGRAAD RFGRFVSWLS TIGITLGVMA LVTVLSVMNG FERELQNNIL GLMPQAILSS EHGSLNPQQL PETAVKLDG VNRVAPITTG DVVLQSARSV AVGVMLGIDP AQKDPLTPYL VNVKQTDLEP GKYNVILGEQ LASQLGVNRG D QIRVMVPS ASQFTPMGRI PSQRLFNVIG TFAANSEVDG YEMLVNIEDA SRLMRYPAGN ITGWRLWLDE PLKVDSLSQQ KL PEGSKWQ DWRDRKGELF QAVRMEKNMM GLLLSLIVAV AAFNIITSLG LMVMEKQGEV AILQTQGLTP RQIMMVFMVQ GAS AGIIGA ILGAALGALL ASQLNNLMPI IGVLLDGAAL PVAIEPLQVI VIALVAMAIA LLSTLYPSWR AAATQPAEAL RYE UniProtKB: Transporter |
-Macromolecule #3: Lipoprotein-releasing system ATP-binding protein LolD
| Macromolecule | Name: Lipoprotein-releasing system ATP-binding protein LolD / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.815721 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KILLQCDNLC KRYQEGSVQT DVLHNVSFSV GEGEMMAIVG SSGSGKSTLL HLLGGLDTPT SGDVIFNGQP MSKLSSAAKA ELRNQKLGF IYQFHHLLPD FTALENVAMP LLIGKKKPAE INSRALEMLK AVGLDHRANH RPSELSGGER QRVAIARALV N NPRLVLAD ...String: KILLQCDNLC KRYQEGSVQT DVLHNVSFSV GEGEMMAIVG SSGSGKSTLL HLLGGLDTPT SGDVIFNGQP MSKLSSAAKA ELRNQKLGF IYQFHHLLPD FTALENVAMP LLIGKKKPAE INSRALEMLK AVGLDHRANH RPSELSGGER QRVAIARALV N NPRLVLAD EPTGNLDARN ADSIFQLLGE LNRLQGTAFL VVTHDLQLAK RMSRQLEMRD GRLTAELSMG RLTAELSM UniProtKB: Lipoprotein-releasing system ATP-binding protein LolD |
-Macromolecule #4: ADP ORTHOVANADATE
| Macromolecule | Name: ADP ORTHOVANADATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: AOV |
|---|---|
| Molecular weight | Theoretical: 544.156 Da |
| Chemical component information | 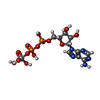 ChemComp-AOV: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 31971 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















