[English] 日本語
 Yorodumi
Yorodumi- EMDB-2372: Negative stain EM structure of the trypsin digested Colicin E9 tr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2372 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
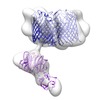
| Title | Negative stain EM structure of the trypsin digested Colicin E9 translocon complex | |||||||||
 Map data Map data | Negative stain EM structure of the trypsin digested Colicin E9 translocon complex comprising OmpF trimer and TolB connected by colicin fragment (residues 2-122). | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Colicins / Porins / Translocon | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to bacteriocin / cell septum assembly / regulation of membrane invagination / bacteriocin transport / extrachromosomal circular DNA / protein import / cell division site / protein transport / outer membrane-bounded periplasmic space / endonuclease activity ...cellular response to bacteriocin / cell septum assembly / regulation of membrane invagination / bacteriocin transport / extrachromosomal circular DNA / protein import / cell division site / protein transport / outer membrane-bounded periplasmic space / endonuclease activity / killing of cells of another organism / Hydrolases; Acting on ester bonds / periplasmic space / defense response to bacterium / protein domain specific binding / cell division / protein-containing complex binding / protein-containing complex / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | Lukoyanova N / Housden NG / Hopper JTS / Rodriguez-Larrea D / Wojdyla JA / Klein A / Kaminska R / Bayley H / Robinson CV / Kleanthous C / Saibil HR | |||||||||
 Citation Citation |  Journal: Science / Year: 2013 Journal: Science / Year: 2013Title: Intrinsically disordered protein threads through the bacterial outer-membrane porin OmpF. Authors: Nicholas G Housden / Jonathan T S Hopper / Natalya Lukoyanova / David Rodriguez-Larrea / Justyna A Wojdyla / Alexander Klein / Renata Kaminska / Hagan Bayley / Helen R Saibil / Carol V ...Authors: Nicholas G Housden / Jonathan T S Hopper / Natalya Lukoyanova / David Rodriguez-Larrea / Justyna A Wojdyla / Alexander Klein / Renata Kaminska / Hagan Bayley / Helen R Saibil / Carol V Robinson / Colin Kleanthous /  Abstract: Porins are β-barrel outer-membrane proteins through which small solutes and metabolites diffuse that are also exploited during cell death. We have studied how the bacteriocin colicin E9 (ColE9) ...Porins are β-barrel outer-membrane proteins through which small solutes and metabolites diffuse that are also exploited during cell death. We have studied how the bacteriocin colicin E9 (ColE9) assembles a cytotoxic translocon at the surface of Escherichia coli that incorporates the trimeric porin OmpF. Formation of the translocon involved ColE9's unstructured N-terminal domain threading in opposite directions through two OmpF subunits, capturing its target TolB on the other side of the membrane in a fixed orientation that triggers colicin import. Thus, an intrinsically disordered protein can tunnel through the narrow pores of an oligomeric porin to deliver an epitope signal to the cell to initiate cell death. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2372.map.gz emd_2372.map.gz | 57.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2372-v30.xml emd-2372-v30.xml emd-2372.xml emd-2372.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2372_image.png EMD-2372_image.png | 143.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2372 http://ftp.pdbj.org/pub/emdb/structures/EMD-2372 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2372 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2372 | HTTPS FTP |
-Validation report
| Summary document |  emd_2372_validation.pdf.gz emd_2372_validation.pdf.gz | 180.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2372_full_validation.pdf.gz emd_2372_full_validation.pdf.gz | 179.4 KB | Display | |
| Data in XML |  emd_2372_validation.xml.gz emd_2372_validation.xml.gz | 6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2372 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2372 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2372 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2372 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2372.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2372.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain EM structure of the trypsin digested Colicin E9 translocon complex comprising OmpF trimer and TolB connected by colicin fragment (residues 2-122). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
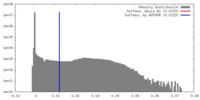
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Trypsin digested colicin E9 translocon complex comprising OmpF tr...
| Entire | Name: Trypsin digested colicin E9 translocon complex comprising OmpF trimer and TolB connected by Colicin fragment (residues 2-122). |
|---|---|
| Components |
|
-Supramolecule #1000: Trypsin digested colicin E9 translocon complex comprising OmpF tr...
| Supramolecule | Name: Trypsin digested colicin E9 translocon complex comprising OmpF trimer and TolB connected by Colicin fragment (residues 2-122). type: sample / ID: 1000 Oligomeric state: OmpF trimer and TolB connected by colicin fragment Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 166.528 KDa / Theoretical: 166.572 KDa / Method: native state ESI-MS |
-Macromolecule #1: Outer membrane porin 1a (Ia;b;F)
| Macromolecule | Name: Outer membrane porin 1a (Ia;b;F) / type: protein_or_peptide / ID: 1 / Name.synonym: OmpF / Number of copies: 1 / Oligomeric state: trimer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 111.294 KDa / Theoretical: 111.253 KDa |
| Sequence | UniProtKB: UNIPROTKB: C4ZQ55 |
-Macromolecule #2: Colicin-E9
| Macromolecule | Name: Colicin-E9 / type: protein_or_peptide / ID: 2 / Name.synonym: A33C colicin E9 residues 2-122 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Colicin-E9 |
-Macromolecule #3: Protein TolB
| Macromolecule | Name: Protein TolB / type: protein_or_peptide / ID: 3 / Name.synonym: P201C TolBHis6 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 44 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Tol-Pal system protein TolB |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 6.5 / Details: 1% (w/v) n-octyl-D-glucopyranoside, 20 mM MES |
| Staining | Type: NEGATIVE Details: Grids with adsorbed protein floated on 2% w/v uranyl acetate for 60 seconds |
| Grid | Details: 300 square mesh copper grid with ~9nm carbon support, negatively glow discharged |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 135,000 times magnification |
| Date | Dec 11, 2012 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN / Digitization - Sampling interval: 15 µm / Number real images: 92 / Average electron dose: 25 e/Å2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 80925 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 0.8 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 80000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Individual images (n=15,500) of the digested translocon complex were interactively selected from micrographs using the EMAN/Boxer software package. Particle images were normalized and band-pass filtered between 150 A and 8 A. Both angular reconstitution sub-tomogram averages were used as initial models for independent projection matching with SPIDER. Independently of initial model for projection matching, final reconstructions displayed similar features at ~20 A resolution estimated by FSC at 0.5 correlation. |
|---|---|
| CTF correction | Details: phases corrected only |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 0.5 CUT-OFF Software - Name: CTFFIND, Spider, EMAN1/Boxer, Imagic, IMOD, Peet Details: Both reconstruction by angular reconstitution and selected sub-tomogram averages were used as initial models for independent projection matching with SPIDER. Independently of initial model ...Details: Both reconstruction by angular reconstitution and selected sub-tomogram averages were used as initial models for independent projection matching with SPIDER. Independently of initial model for projection matching, final reconstructions displayed similar features at ~20 A resolution estimated by FSC at 0.5 correlation Number images used: 7000 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | crystal structures were manually fitted into the EM map |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | crystal structures were manually fitted into the EM map |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)