+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4061 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM map of E. coli BAM complex (BamABCDE) in DDM micelle | |||||||||
 Map data Map data | E. coli BamABCDE | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationBam protein complex / Gram-negative-bacterium-type cell outer membrane assembly / Secretion of toxins / protein insertion into membrane / cell outer membrane / protein-macromolecule adaptor activity / cell adhesion / response to antibiotic / cell surface / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
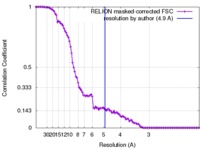
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Iadanza MG / Ranson NA / Radford SE / Higgins AJ / Schriffin B / Calabrase AN / Ashcroft AE / Brockwell DJ | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2016 Journal: Nat Commun / Year: 2016Title: Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Authors: Matthew G Iadanza / Anna J Higgins / Bob Schiffrin / Antonio N Calabrese / David J Brockwell / Alison E Ashcroft / Sheena E Radford / Neil A Ranson /  Abstract: The β-barrel assembly machinery (BAM) is a ∼203 kDa complex of five proteins (BamA-E), which is essential for viability in E. coli. BAM promotes the folding and insertion of β-barrel proteins ...The β-barrel assembly machinery (BAM) is a ∼203 kDa complex of five proteins (BamA-E), which is essential for viability in E. coli. BAM promotes the folding and insertion of β-barrel proteins into the outer membrane via a poorly understood mechanism. Several current models suggest that BAM functions through a 'lateral gating' motion of the β-barrel of BamA. Here we present a cryo-EM structure of the BamABCDE complex, at 4.9 Å resolution. The structure is in a laterally open conformation showing that gating is independent of BamB binding. We describe conformational changes throughout the complex and interactions between BamA, B, D and E, and the detergent micelle that suggest communication between BAM and the lipid bilayer. Finally, using an enhanced reconstitution protocol and functional assays, we show that for the outer membrane protein OmpT, efficient folding in vitro requires lateral gating in BAM. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4061.map.gz emd_4061.map.gz | 5.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4061-v30.xml emd-4061-v30.xml emd-4061.xml emd-4061.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4061_fsc.xml emd_4061_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_4061.png emd_4061.png | 117.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4061 http://ftp.pdbj.org/pub/emdb/structures/EMD-4061 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4061 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4061 | HTTPS FTP |
-Related structure data
| Related structure data |  5ljoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4061.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4061.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | E. coli BamABCDE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : BAM complex
| Entire | Name: BAM complex |
|---|---|
| Components |
|
-Supramolecule #1: BAM complex
| Supramolecule | Name: BAM complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 200 KDa |
-Macromolecule #1: BamA
| Macromolecule | Name: BamA / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MAMKKLLIAS LLFSSATVYG AEGFVVKDIH FEGLQRVAVG AALLSMPVRT GDTVNDEDIS NTIRALFAT GNFEDVRVLR DGDTLLVQVK ERPTIASITF SGNKSVKDDM LKQNLEASGV R VGESLDRT TIADIEKGLE DFYYSVGKYS ASVKAVVTPL PRNRVDLKLV ...String: MAMKKLLIAS LLFSSATVYG AEGFVVKDIH FEGLQRVAVG AALLSMPVRT GDTVNDEDIS NTIRALFAT GNFEDVRVLR DGDTLLVQVK ERPTIASITF SGNKSVKDDM LKQNLEASGV R VGESLDRT TIADIEKGLE DFYYSVGKYS ASVKAVVTPL PRNRVDLKLV FQEGVSAEIQ QI NIVGNHA FTTDELISHF QLRDEVPWWN VVGDRKYQKQ KLAGDLETLR SYYLDRGYAR FNI DSTQVS LTPDKKGIYV TVNITEGDQY KLSGVEVSGN LAGHSAEIEQ LTKIEPGELY NGTK VTKME DDIKKLLGRY GYAYPRVQSM PEINDADKTV KLRVNVDAGN RFYVRKIRFE GNDTS KDAV LRREMRQMEG AWLGSDLVDQ GKERLNRLGF FETVDTDTQR VPGSPDQVDV VYKVKE RNT GSFNFGIGYG TESGVSFQAG VQQDNWLGTG YAVGINGTKN DYQTYAELSV TNPYFTV DG VSLGGRLFYN DFQADDADLS DYTNKSYGTD VTLGFPINEY NSLRAGLGYV HNSLSNMQ P QVAMWRYLYS MGEHPSTSDQ DNSFKTDDFT FNYGWTYNKL DRGYFPTDGS RVNLTGKVT IPGSDNEYYK VTLDTATYVP IDDDHKWVVL GRTRWGYGDG LGGKEMPFYE NFYAGGSSTV RGFQSNTIG PKAVYFPHQA SNYDPDYDYE CATQDGAKDL CKSDDAVGGN AMAVASLEFI T PTPFISDK YANSVRTSFF WDMGTVWDTN WDSSQYSGYP DYSDPSNIRM SAGIALQWMS PL GPLVFSY AQPFKKYDGD KAEQFQFNIG KTW |
-Macromolecule #2: BamB
| Macromolecule | Name: BamB / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MQLRKLLLPG LLSVTLLSGC SLFNSEEDVV KMSPLPTVEN QFTPTTAWST SVGSGIGNFY SNLHPALAD NVVYAADRAG LVKALNADDG KEIWSVSLAE KDGWFSKEPA LLSGGVTVSG G HVYIGSEK AQVYALNTSD GTVAWQTKVA GEALSRPVVS DGLVLIHTSN ...String: MQLRKLLLPG LLSVTLLSGC SLFNSEEDVV KMSPLPTVEN QFTPTTAWST SVGSGIGNFY SNLHPALAD NVVYAADRAG LVKALNADDG KEIWSVSLAE KDGWFSKEPA LLSGGVTVSG G HVYIGSEK AQVYALNTSD GTVAWQTKVA GEALSRPVVS DGLVLIHTSN GQLQALNEAD GA VKWTVNL DMPSLSLRGE SAPTTAFGAA VVGGDNGRVS AVLMEQGQMI WQQRISQATG STE IDRLSD VDTTPVVVNG VVFALAYNGN LTALDLRSGQ IMWKRELGSV NDFIVDGNRI YLVD QNDRV MALTIDGGVT LWTQSDLLHR LLTSPVLYNG NLVVGDSEGY LHWINVEDGR FVAQQ KVDS SGFQTEPVAA DGKLLIQAKD GTVYSITR |
-Macromolecule #3: BamC
| Macromolecule | Name: BamC / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MAYSVQKSRL AKVAGVSLVL LLAACSSDSR YKRQVSGDEA YLEAAPLAEL HAPAGMILPV TSGDYAIPV TNGSGAVGKA LDIRPPAQPL ALVSGARTQF TGDTASLLVE NGRGNTLWPQ V VSVLQAKN YTITQRDDAG QTLTTDWVQW NRLDEDEQYR GRYQISVKPQ ...String: MAYSVQKSRL AKVAGVSLVL LLAACSSDSR YKRQVSGDEA YLEAAPLAEL HAPAGMILPV TSGDYAIPV TNGSGAVGKA LDIRPPAQPL ALVSGARTQF TGDTASLLVE NGRGNTLWPQ V VSVLQAKN YTITQRDDAG QTLTTDWVQW NRLDEDEQYR GRYQISVKPQ GYQQAVTVKL LN LEQAGKP VADAASMQRY STEMMNVISA GLDKSATDAA NAAQNRASTT MDVQSAADDT GLP MLVVRG PFNVVWQRLP AALEKVGMKV TDSTRSQGNM AVTYKPLSDS DWQELGASDP GLAS GDYKL QVGDLDNRSS LQFIDPKGHT LTQSQNDALV AVFQAAFSK |
-Macromolecule #4: BamD
| Macromolecule | Name: BamD / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MTRMKYLVAA ATLSLFLAGC SGSKEEVPDN PPNEIYATAQ QKLQDGNWRQ AITQLEALDN RYPFGPYSQ QVQLDLIYAY YKNADLPLAQ AAIDRFIRLN PTHPNIDYVM YMRGLTNMAL D DSALQGFF GVDRSDRDPQ HARAAFSDFS KLVRGYPNSQ YTTDATKRLV ...String: MTRMKYLVAA ATLSLFLAGC SGSKEEVPDN PPNEIYATAQ QKLQDGNWRQ AITQLEALDN RYPFGPYSQ QVQLDLIYAY YKNADLPLAQ AAIDRFIRLN PTHPNIDYVM YMRGLTNMAL D DSALQGFF GVDRSDRDPQ HARAAFSDFS KLVRGYPNSQ YTTDATKRLV FLKDRLAKYE YS VAEYYTE RGAWVAVVNR VEGMLRDYPD TQATRDALPL MENAYRQMQM NAQAEKVAKI IAA NSSNT |
-Macromolecule #5: BamE
| Macromolecule | Name: BamE / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MRCKTLTAAA AVLLMLTAGC STLERVVYRP DINQGNYLTA NDVSKIRVGM TQQQVAYALG TPLMSDPFG TNTWFYVFRQ QPGHEGVTQQ TLTLTFNSSG VLTNIDNKPA LSGN |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R3.5/1 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa / Details: Pelco EasyGlo | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: LEICA EM GP Details: Double Blotted: 3ul sample applied, hand blotted then additional 3ul sample applied and blotted and plunge frozen. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Digitization - Dimensions - Width: 3840 pixel / Digitization - Dimensions - Height: 3712 pixel / Digitization - Sampling interval: 5.0 µm / Number grids imaged: 1 / Number real images: 7204 / Average exposure time: 10.0 sec. / Average electron dose: 40.0 e/Å2 Details: Collected in movie mode at 2 FPS Final data used frames 4-14 for total dose of 20 electrons per square Angstrom |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.5 µm / Calibrated defocus min: 1.5 µm / Calibrated magnification: 48076 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.6 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 48076 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-5ljo: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)