[English] 日本語
 Yorodumi
Yorodumi- EMDB-23665: Structural basis for SARS-CoV-2 envelope protein in recognition o... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23665 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural basis for SARS-CoV-2 envelope protein in recognition of human cell junction protein PALS1 | |||||||||
 Map data Map data | Cryosparc non-uniform refinement without sharpening | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 envelope protein / PDZ-binding motif / complex / pathogen-host interaction / CELL ADHESION-VIRAL PROTEIN complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein localization to myelin sheath abaxonal region / disruption of cellular anatomical structure in another organism / SARS-CoV-1 targets PDZ proteins in cell-cell junction / symbiont-mediated perturbation of host cell endomembrane system / viral budding from Golgi membrane / myelin assembly / establishment or maintenance of polarity of embryonic epithelium / morphogenesis of an epithelial sheet / Tight junction interactions / myelin sheath adaxonal region ...protein localization to myelin sheath abaxonal region / disruption of cellular anatomical structure in another organism / SARS-CoV-1 targets PDZ proteins in cell-cell junction / symbiont-mediated perturbation of host cell endomembrane system / viral budding from Golgi membrane / myelin assembly / establishment or maintenance of polarity of embryonic epithelium / morphogenesis of an epithelial sheet / Tight junction interactions / myelin sheath adaxonal region / SARS-CoV-2 targets PDZ proteins in cell-cell junction / regulation of transforming growth factor beta receptor signaling pathway / Regulation of gap junction activity / lateral loop / Schmidt-Lanterman incisure / establishment or maintenance of epithelial cell apical/basal polarity / peripheral nervous system myelin maintenance / generation of neurons / apical junction complex / central nervous system neuron development / host cell Golgi membrane / bicellular tight junction / endoplasmic reticulum-Golgi intermediate compartment membrane / Maturation of protein E / protein localization to plasma membrane / adherens junction / cerebral cortex development / gene expression / Translation of Structural Proteins / Virion Assembly and Release / Induction of Cell-Cell Fusion / perikaryon / Attachment and Entry / apical plasma membrane / protein domain specific binding / axon / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / Golgi apparatus / protein-containing complex / extracellular exosome / ATP binding / identical protein binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
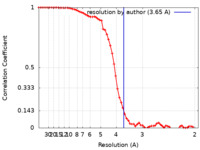
| Method | single particle reconstruction / cryo EM / Resolution: 3.65 Å | |||||||||
 Authors Authors | Liu Q / Chai J | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural basis for SARS-CoV-2 envelope protein recognition of human cell junction protein PALS1. Authors: Jin Chai / Yuanheng Cai / Changxu Pang / Liguo Wang / Sean McSweeney / John Shanklin / Qun Liu /  Abstract: The COVID-19 pandemic, caused by the SARS-CoV-2 virus, has created global health and economic emergencies. SARS-CoV-2 viruses promote their own spread and virulence by hijacking human proteins, which ...The COVID-19 pandemic, caused by the SARS-CoV-2 virus, has created global health and economic emergencies. SARS-CoV-2 viruses promote their own spread and virulence by hijacking human proteins, which occurs through viral protein recognition of human targets. To understand the structural basis for SARS-CoV-2 viral-host protein recognition, here we use cryo-electron microscopy (cryo-EM) to determine a complex structure of the human cell junction protein PALS1 and SARS-CoV-2 viral envelope (E) protein. Our reported structure shows that the E protein C-terminal DLLV motif recognizes a pocket formed exclusively by hydrophobic residues from the PDZ and SH3 domains of PALS1. Our structural analysis provides an explanation for the observation that the viral E protein recruits PALS1 from lung epithelial cell junctions. In addition, our structure provides novel targets for peptide- and small-molecule inhibitors that could block the PALS1-E interactions to reduce E-mediated virulence. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23665.map.gz emd_23665.map.gz | 31.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23665-v30.xml emd-23665-v30.xml emd-23665.xml emd-23665.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23665_fsc.xml emd_23665_fsc.xml | 6.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_23665.png emd_23665.png | 152.4 KB | ||
| Filedesc metadata |  emd-23665.cif.gz emd-23665.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23665 http://ftp.pdbj.org/pub/emdb/structures/EMD-23665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23665 | HTTPS FTP |
-Related structure data
| Related structure data |  7m4rMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23665.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23665.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc non-uniform refinement without sharpening | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.684 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex structure of SARS-CoV-2 envelope protein Ec18 and human P...
| Entire | Name: Complex structure of SARS-CoV-2 envelope protein Ec18 and human PALS1 PSG domains |
|---|---|
| Components |
|
-Supramolecule #1: Complex structure of SARS-CoV-2 envelope protein Ec18 and human P...
| Supramolecule | Name: Complex structure of SARS-CoV-2 envelope protein Ec18 and human PALS1 PSG domains type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: MAGUK p55 subfamily member 5
| Macromolecule | Name: MAGUK p55 subfamily member 5 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.780504 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAPITDERV YESIGQYGGE TVKIVRIEKA RDIPLGATVR NEMDSVIISR IVKGGAAEKS GLLHEGDEVL EINGIEIRGK DVNEVFDLL SDMHGTLTFV LIPSQQIKPP PAKETVIHVK AHFDYDPSDD PYVPCRELGL SFQKGDILHV ISQEDPNWWQ A YREGDEDN ...String: SNAPITDERV YESIGQYGGE TVKIVRIEKA RDIPLGATVR NEMDSVIISR IVKGGAAEKS GLLHEGDEVL EINGIEIRGK DVNEVFDLL SDMHGTLTFV LIPSQQIKPP PAKETVIHVK AHFDYDPSDD PYVPCRELGL SFQKGDILHV ISQEDPNWWQ A YREGDEDN QPLAGLVPGK EEILTYEEMS LYHQPANRKR PIILIGPQNC GQNELRQRLM NKEKDRFASA VPHTTRSRRD QE VAGRDYH FVSRQAFEAD IAAGKFIEHG EFEKNLYGTS IDSVRQVINS GKICLLSLRT QSLKTLRNSD LKPYIIFIAP PSQ ERLRAL LAKEGKNPKP EELREIIEKT REMEQNNGHY FDTAIVNSDL DKAYQELLRL INKLDTEPQW VPSTWLR UniProtKB: Protein PALS1, Protein PALS1 |
-Macromolecule #2: Envelope small membrane protein
| Macromolecule | Name: Envelope small membrane protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.062395 KDa |
| Sequence | String: VYSRVKNLNS SRVPDLLV UniProtKB: Envelope small membrane protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)