[English] 日本語
 Yorodumi
Yorodumi- EMDB-23587: Cryo-EM structure of the elongation module of the bacillamide NRP... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23587 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
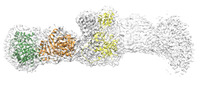
| Title | Cryo-EM structure of the elongation module of the bacillamide NRPS, BmdB, in complex with the oxidase, BmdC | |||||||||
 Map data Map data | EM map of the locally refined bacillamide NRPS, BmdB, complexed with the oxidase, BmdC | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nonribosomal peptide synthetases Oxidase Bacillamide synthetases / BIOSYNTHETIC PROTEIN | |||||||||
| Biological species |  Thermoactinomyces vulgaris (bacteria) Thermoactinomyces vulgaris (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Sharon I / Strauss M / Schmeing TM | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module. Authors: Camille Marie Fortinez / Kristjan Bloudoff / Connor Harrigan / Itai Sharon / Mike Strauss / T Martin Schmeing /  Abstract: Nonribosomal peptide synthetases (NRPSs) are large modular enzymes that synthesize secondary metabolites and natural product therapeutics. Most NRPS biosynthetic pathways include an NRPS and ...Nonribosomal peptide synthetases (NRPSs) are large modular enzymes that synthesize secondary metabolites and natural product therapeutics. Most NRPS biosynthetic pathways include an NRPS and additional proteins that introduce chemical modifications before, during or after assembly-line synthesis. The bacillamide biosynthetic pathway is a common, three-protein system, with a decarboxylase that prepares an NRPS substrate, an NRPS, and an oxidase. Here, the pathway is reconstituted in vitro. The oxidase is shown to perform dehydrogenation of the thiazoline in the peptide intermediate while it is covalently attached to the NRPS, as the penultimate step in bacillamide D synthesis. Structural analysis of the oxidase reveals a dimeric, two-lobed architecture with a remnant RiPP recognition element and a dramatic wrapping loop. The oxidase forms a stable complex with the NRPS and dimerizes it. We visualized co-complexes of the oxidase bound to the elongation module of the NRPS using X-ray crystallography and cryo-EM. The three active sites (for adenylation, condensation/cyclization, and oxidation) form an elegant arc to facilitate substrate delivery. The structures enabled a proof-of-principle bioengineering experiment in which the BmdC oxidase domain is embedded into the NRPS. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23587.map.gz emd_23587.map.gz | 398.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23587-v30.xml emd-23587-v30.xml emd-23587.xml emd-23587.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23587_fsc.xml emd_23587_fsc.xml | 16.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_23587.png emd_23587.png | 236.7 KB | ||
| Masks |  emd_23587_msk_1.map emd_23587_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23587.cif.gz emd-23587.cif.gz | 6 KB | ||
| Others |  emd_23587_half_map_1.map.gz emd_23587_half_map_1.map.gz emd_23587_half_map_2.map.gz emd_23587_half_map_2.map.gz | 391.9 MB 391.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23587 http://ftp.pdbj.org/pub/emdb/structures/EMD-23587 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23587 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23587 | HTTPS FTP |
-Related structure data
| Related structure data |  7ly4MC  7ly5C  7ly6C  7ly7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23587.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23587.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map of the locally refined bacillamide NRPS, BmdB, complexed with the oxidase, BmdC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.14 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23587_msk_1.map emd_23587_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A of the locally refined bacillamide...
| File | emd_23587_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of the locally refined bacillamide NRPS, BmdB, complexed with the oxidase, BmdC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B of the locally refined bacillamide...
| File | emd_23587_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of the locally refined bacillamide NRPS, BmdB, complexed with the oxidase, BmdC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Elongation module of the bacillamide NRPS, BmdB, in complex with ...
| Entire | Name: Elongation module of the bacillamide NRPS, BmdB, in complex with the oxidase, BmdC |
|---|---|
| Components |
|
-Supramolecule #1: Elongation module of the bacillamide NRPS, BmdB, in complex with ...
| Supramolecule | Name: Elongation module of the bacillamide NRPS, BmdB, in complex with the oxidase, BmdC type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Thermoactinomyces vulgaris (bacteria) Thermoactinomyces vulgaris (bacteria) |
-Macromolecule #1: BmdC, Oxidase
| Macromolecule | Name: BmdC, Oxidase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermoactinomyces vulgaris (bacteria) Thermoactinomyces vulgaris (bacteria) |
| Molecular weight | Theoretical: 38.177254 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAMGEQIFYW SPAKHWRMSD EGVVIGESTY TGMILEWFPE FYFFAQTGVT INRLLERFSS GSEKEANEIL ELLIQDRVLV EGILPPREV FSPQEGLFVN PYSEQIRYSK EALDYYVSEQ LNRTHAACRS TKIQLETSGA LPDIIQKRRS CRRFDMKTPV S FATFSNLL ...String: GAMGEQIFYW SPAKHWRMSD EGVVIGESTY TGMILEWFPE FYFFAQTGVT INRLLERFSS GSEKEANEIL ELLIQDRVLV EGILPPREV FSPQEGLFVN PYSEQIRYSK EALDYYVSEQ LNRTHAACRS TKIQLETSGA LPDIIQKRRS CRRFDMKTPV S FATFSNLL SSLKQRKEDK ILYNYASAGG LYPIDVFVYV KPRRVEGVKA GFYYFNPADH SLVLVNNIDQ VIKDDHELIN QD IFAQSAF SVYLVYNARA SMPKYGAAGY FYACIEAGII TATLNMVAED LNVGLCSIGH MNFEEIQTFL KLEDHQVILH AIE GGLKID GAAAENLYFQ |
-Macromolecule #2: BmdB, bacillamide NRPS
| Macromolecule | Name: BmdB, bacillamide NRPS / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermoactinomyces vulgaris (bacteria) Thermoactinomyces vulgaris (bacteria) |
| Molecular weight | Theoretical: 117.373578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAMEADLSEP FSLTEVQTAY MLGRNPQFEL SGISPQTYFE YETELDIARL SRSFQKVIQR HPMLRAVILP EGKQQILRDV PEYEIEVES LVSMPPEKQA ARLREERSRM IDHVFPLGQW PLFELKAFQL QEHTYLLCFR YDALLMDGAS MNLVGQDLMH Y YHQPDAQL ...String: GAMEADLSEP FSLTEVQTAY MLGRNPQFEL SGISPQTYFE YETELDIARL SRSFQKVIQR HPMLRAVILP EGKQQILRDV PEYEIEVES LVSMPPEKQA ARLREERSRM IDHVFPLGQW PLFELKAFQL QEHTYLLCFR YDALLMDGAS MNLVGQDLMH Y YHQPDAQL PPLSFTFQDY MHIYDDMKRG TEYETAKAYW TNKLPDFPPA PSLLLAKDPA EIGTPNFQSL TTIITKDKWL KL RRLAQDK QVTPSALLCT VYGEVLAFWS NQRRLAINLT VFNRYPVHDE VEQIVGDFTS LILLDMDMDQ KQPFFTKVEQ TQS TLLDGL EHRHYDGVEF IRDYTRYHQM RPKAVMPIVF TSMLAGAGAF AWEEIGSLRH IHARTPQVYL DNVVIEKNGE LLVS WNYVE ELFDAEVMES MFTQFVELLD QLVEQGDINP LRISQKDYAL IDQYNATAEP IPAATLHQLF IDQAQRTPDQ VAVVF EQEW LTYSELDQRS NQVARFLQSR GIGRGDRVGV LAKRQVETII NLMAVLKAGA AYVPIDPDHP YERQTYILEN SSCKIL LDS DLYETMEISS YADGDLTPVA EPEDTAYVIY TSGSTGRPKG VIITHQAASN TIQDINRKFE VNEEDRIIGI SSMCFDL SV YDIFGTLSAG ATLVMIRDPR DMRELVRTVE RRGITIWNTV PAIMDLALDH VGSHFENISL RLVLLSGDWI PLPLPAKI N RHFPVADVIS LGGATEASIW SIYWPIEQVE ANWKSIPYGK PLANQTYYVL NYDQKMCPVG VIGDLYIGGA GLAQGYLND DQKTKDAFIM HPEFGPIYKT GDCGRMRPEG YIEFLGRQDY QVKIQGYRVE LEEISHCLLT YPDVDQAVVI DQTDERGMKF LVGYVVAQQ EIDEKALRKH LMEHLPEYMI PAHLVHLEQL PLTPNGKLDR KALPVPKKQR NAEKFVAPQA GLEKILASVW Q EVLNVEQI GANDHFFALG GDSIKAIQVS ARLFVQGYHL DTKSLFEFPV LRDVARTIKK LAAAENLYFQ |
-Macromolecule #3: FLAVIN MONONUCLEOTIDE
| Macromolecule | Name: FLAVIN MONONUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 2 / Formula: FMN |
|---|---|
| Molecular weight | Theoretical: 456.344 Da |
| Chemical component information |  ChemComp-FMN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 109.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)