+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23509 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | NHEJ Short-range synaptic complex | |||||||||||||||||||||
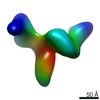
 Map data Map data | Synaptic complex | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | NHEJ / DNA BINDING PROTEIN / DNA BINDING PROTEIN-DNA complex | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationFHA domain binding / positive regulation of chromosome organization / positive regulation of ligase activity / DNA ligase IV complex / DNA ligase activity / Ku70:Ku80 complex / DN2 thymocyte differentiation / negative regulation of t-circle formation / DNA ligase (ATP) / T cell receptor V(D)J recombination ...FHA domain binding / positive regulation of chromosome organization / positive regulation of ligase activity / DNA ligase IV complex / DNA ligase activity / Ku70:Ku80 complex / DN2 thymocyte differentiation / negative regulation of t-circle formation / DNA ligase (ATP) / T cell receptor V(D)J recombination / DNA end binding / pro-B cell differentiation / small-subunit processome assembly / positive regulation of lymphocyte differentiation / DNA ligase (ATP) activity / DNA-dependent protein kinase complex / DNA-dependent protein kinase-DNA ligase 4 complex / nonhomologous end joining complex / immunoglobulin V(D)J recombination / nucleotide-excision repair, DNA gap filling / single strand break repair / regulation of smooth muscle cell proliferation / cellular response to X-ray / V(D)J recombination / nuclear telomere cap complex / double-strand break repair via classical nonhomologous end joining / protein localization to site of double-strand break / isotype switching / Cytosolic sensors of pathogen-associated DNA / IRF3-mediated induction of type I IFN / positive regulation of neurogenesis / regulation of telomere maintenance / recombinational repair / U3 snoRNA binding / protein localization to chromosome, telomeric region / DNA biosynthetic process / response to ionizing radiation / cellular response to lithium ion / cellular hyperosmotic salinity response / 2-LTR circle formation / telomeric DNA binding / hematopoietic stem cell proliferation / ligase activity / positive regulation of protein kinase activity / site of DNA damage / Lyases; Carbon-oxygen lyases; Other carbon-oxygen lyases / T cell differentiation / somatic stem cell population maintenance / 5'-deoxyribose-5-phosphate lyase activity / response to X-ray / hematopoietic stem cell differentiation / ATP-dependent activity, acting on DNA / chromosome organization / telomere maintenance via telomerase / SUMOylation of DNA damage response and repair proteins / condensed chromosome / DNA polymerase binding / neurogenesis / cellular response to ionizing radiation / telomere maintenance / activation of innate immune response / DNA helicase activity / cyclin binding / cellular response to leukemia inhibitory factor / B cell differentiation / central nervous system development / stem cell proliferation / response to gamma radiation / small-subunit processome / Nonhomologous End-Joining (NHEJ) / enzyme activator activity / cellular response to gamma radiation / protein-DNA complex / base-excision repair / double-strand break repair via nonhomologous end joining / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / establishment of integrated proviral latency / fibrillar center / positive regulation of fibroblast proliferation / site of double-strand break / double-strand break repair / T cell differentiation in thymus / double-stranded DNA binding / neuron apoptotic process / scaffold protein binding / fibroblast proliferation / secretory granule lumen / DNA recombination / transcription regulator complex / damaged DNA binding / in utero embryonic development / ficolin-1-rich granule lumen / negative regulation of neuron apoptotic process / chromosome, telomeric region / cell population proliferation / transcription cis-regulatory region binding / ribonucleoprotein complex / innate immune response / cell division / negative regulation of DNA-templated transcription Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.4 Å | |||||||||||||||||||||
 Authors Authors | He Y / Chen S | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structural basis of long-range to short-range synaptic transition in NHEJ. Authors: Siyu Chen / Linda Lee / Tasmin Naila / Susan Fishbain / Annie Wang / Alan E Tomkinson / Susan P Lees-Miller / Yuan He /   Abstract: DNA double-strand breaks (DSBs) are a highly cytotoxic form of DNA damage and the incorrect repair of DSBs is linked to carcinogenesis. The conserved error-prone non-homologous end joining (NHEJ) ...DNA double-strand breaks (DSBs) are a highly cytotoxic form of DNA damage and the incorrect repair of DSBs is linked to carcinogenesis. The conserved error-prone non-homologous end joining (NHEJ) pathway has a key role in determining the effects of DSB-inducing agents that are used to treat cancer as well as the generation of the diversity in antibodies and T cell receptors. Here we applied single-particle cryo-electron microscopy to visualize two key DNA-protein complexes that are formed by human NHEJ factors. The Ku70/80 heterodimer (Ku), the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs), DNA ligase IV (LigIV), XRCC4 and XLF form a long-range synaptic complex, in which the DNA ends are held approximately 115 Å apart. Two DNA end-bound subcomplexes comprising Ku and DNA-PKcs are linked by interactions between the DNA-PKcs subunits and a scaffold comprising LigIV, XRCC4, XLF, XRCC4 and LigIV. The relative orientation of the DNA-PKcs molecules suggests a mechanism for autophosphorylation in trans, which leads to the dissociation of DNA-PKcs and the transition into the short-range synaptic complex. Within this complex, the Ku-bound DNA ends are aligned for processing and ligation by the XLF-anchored scaffold, and a single catalytic domain of LigIV is stably associated with a nick between the two Ku molecules, which suggests that the joining of both strands of a DSB involves both LigIV molecules. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23509.map.gz emd_23509.map.gz | 23.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23509-v30.xml emd-23509-v30.xml emd-23509.xml emd-23509.xml | 26.8 KB 26.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23509.png emd_23509.png | 104.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23509 http://ftp.pdbj.org/pub/emdb/structures/EMD-23509 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23509 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23509 | HTTPS FTP |
-Validation report
| Summary document |  emd_23509_validation.pdf.gz emd_23509_validation.pdf.gz | 392.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23509_full_validation.pdf.gz emd_23509_full_validation.pdf.gz | 392 KB | Display | |
| Data in XML |  emd_23509_validation.xml.gz emd_23509_validation.xml.gz | 5.6 KB | Display | |
| Data in CIF |  emd_23509_validation.cif.gz emd_23509_validation.cif.gz | 6.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23509 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23509 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23509 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23509 | HTTPS FTP |
-Related structure data
| Related structure data |  7lsyMC  7lt3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23509.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23509.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Synaptic complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.66 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Short-range synaptic complex of NHEJ
+Supramolecule #1: Short-range synaptic complex of NHEJ
+Macromolecule #1: X-ray repair cross-complementing protein 6
+Macromolecule #2: X-ray repair cross-complementing protein 5
+Macromolecule #5: DNA repair protein XRCC4
+Macromolecule #6: Non-homologous end-joining factor 1
+Macromolecule #10: DNA ligase 4
+Macromolecule #3: DNA (26-MER)
+Macromolecule #4: DNA (5'-D(P*GP*TP*TP*CP*TP*TP*AP*GP*TP*AP*TP*AP*TP*A)-3')
+Macromolecule #7: DNA (5'-D(P*TP*AP*TP*AP*TP*AP*CP*TP*AP*AP*GP*AP*AP*C)-3')
+Macromolecule #8: DNA (26-MER)
+Macromolecule #9: DNA (5'-D(P*CP*AP*AP*TP*GP*AP*AP*AP*CP*GP*GP*AP*AP*CP*AP*GP*TP*CP...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R3.5/1 / Material: COPPER / Mesh: 200 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Support film - Film thickness: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 11520 pixel / Digitization - Dimensions - Height: 8184 pixel / Number grids imaged: 2 / Number real images: 32723 / Average exposure time: 0.0426 sec. / Average electron dose: 46.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 30000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-7lsy: |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















 unidentified baculovirus
unidentified baculovirus
