[English] 日本語
 Yorodumi
Yorodumi- EMDB-23351: Structure of Arabidopsis thaliana sulfate transporter AtSULTR4;1 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23351 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Arabidopsis thaliana sulfate transporter AtSULTR4;1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Sulfate transport / SLC26 / MEMBRANE PROTEIN / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationchloroplast membrane / secondary active sulfate transmembrane transporter activity / symporter activity Similarity search - Function | |||||||||
| Biological species |  | |||||||||
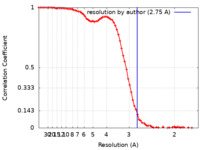
| Method | single particle reconstruction / cryo EM / Resolution: 2.75 Å | |||||||||
 Authors Authors | Wang L / Chen K | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structure and function of an Arabidopsis thaliana sulfate transporter. Authors: Lie Wang / Kehan Chen / Ming Zhou /  Abstract: Plant sulfate transporters (SULTR) mediate absorption and distribution of sulfate (SO) and are essential for plant growth; however, our understanding of their structures and functions remains ...Plant sulfate transporters (SULTR) mediate absorption and distribution of sulfate (SO) and are essential for plant growth; however, our understanding of their structures and functions remains inadequate. Here we present the structure of a SULTR from Arabidopsis thaliana, AtSULTR4;1, in complex with SO at an overall resolution of 2.8 Å. AtSULTR4;1 forms a homodimer and has a structural fold typical of the SLC26 family of anion transporters. The bound SO is coordinated by side-chain hydroxyls and backbone amides, and further stabilized electrostatically by the conserved Arg393 and two helix dipoles. Proton and SO are co-transported by AtSULTR4;1 and a proton gradient significantly enhances SO transport. Glu347, which is ~7 Å from the bound SO, is required for H-driven transport. The cytosolic STAS domain interacts with transmembrane domains, and deletion of the STAS domain or mutations to the interface compromises dimer formation and reduces SO transport, suggesting a regulatory function of the STAS domain. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23351.map.gz emd_23351.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23351-v30.xml emd-23351-v30.xml emd-23351.xml emd-23351.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23351_fsc.xml emd_23351_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_23351.png emd_23351.png | 295.5 KB | ||
| Filedesc metadata |  emd-23351.cif.gz emd-23351.cif.gz | 6.3 KB | ||
| Others |  emd_23351_half_map_1.map.gz emd_23351_half_map_1.map.gz emd_23351_half_map_2.map.gz emd_23351_half_map_2.map.gz | 28.2 MB 28.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23351 http://ftp.pdbj.org/pub/emdb/structures/EMD-23351 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23351 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23351 | HTTPS FTP |
-Validation report
| Summary document |  emd_23351_validation.pdf.gz emd_23351_validation.pdf.gz | 795.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23351_full_validation.pdf.gz emd_23351_full_validation.pdf.gz | 794.6 KB | Display | |
| Data in XML |  emd_23351_validation.xml.gz emd_23351_validation.xml.gz | 14.1 KB | Display | |
| Data in CIF |  emd_23351_validation.cif.gz emd_23351_validation.cif.gz | 17.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23351 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23351 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23351 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23351 | HTTPS FTP |
-Related structure data
| Related structure data |  7lhvMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23351.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23351.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_23351_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_23351_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Sulfate transporter 4;1
| Entire | Name: Sulfate transporter 4;1 |
|---|---|
| Components |
|
-Supramolecule #1: Sulfate transporter 4;1
| Supramolecule | Name: Sulfate transporter 4;1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 70 KDa |
-Macromolecule #1: Sulfate transporter 4.1, chloroplastic
| Macromolecule | Name: Sulfate transporter 4.1, chloroplastic / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 75.168164 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSYASLSVKD LTSLVSRSGT GSSSSLKPPG QTRPVKVIPL QHPDTSNEAR PPSIPFDDIF SGWTAKIKRM RLVDWIDTLF PCFRWIRTY RWSEYFKLDL MAGITVGIML VPQAMSYAKL AGLPPIYGLY SSFVPVFVYA IFGSSRQLAI GPVALVSLLV S NALGGIAD ...String: MSYASLSVKD LTSLVSRSGT GSSSSLKPPG QTRPVKVIPL QHPDTSNEAR PPSIPFDDIF SGWTAKIKRM RLVDWIDTLF PCFRWIRTY RWSEYFKLDL MAGITVGIML VPQAMSYAKL AGLPPIYGLY SSFVPVFVYA IFGSSRQLAI GPVALVSLLV S NALGGIAD TNEELHIELA ILLALLVGIL ECIMGLLRLG WLIRFISHSV ISGFTSASAI VIGLSQIKYF LGYSIARSSK IV PIVESII AGADKFQWPP FVMGSLILVI LQVMKHVGKA KKELQFLRAA APLTGIVLGT TIAKVFHPPS ISLVGEIPQG LPT FSFPRS FDHAKTLLPT SALITGVAIL ESVGIAKALA AKNRYELDSN SELFGLGVAN ILGSLFSAYP ATGSFSRSAV NNES EAKTG LSGLITGIII GCSLLFLTPM FKYIPQCALA AIVISAVSGL VDYDEAIFLW RVDKRDFSLW TITSTITLFF GIEIG VLVG VGFSLAFVIH ESANPHIAVL GRLPGTTVYR NIKQYPEAYT YNGIVIVRID SPIYFANISY IKDRLREYEV AVDKYT NRG LEVDRINFVI LEMSPVTHID SSAVEALKEL YQEYKTRDIQ LAISNPNKDV HLTIARSGMV ELVGKEWFFV RVHDAVQ VC LQYVQSSNLE DKHLSFTRRY GGSNNNSSSS NALLKEPLLS VEK UniProtKB: Sulfate transporter 4.1, chloroplastic |
-Macromolecule #2: (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate
| Macromolecule | Name: (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate type: ligand / ID: 2 / Number of copies: 4 / Formula: S1P |
|---|---|
| Molecular weight | Theoretical: 379.472 Da |
| Chemical component information | 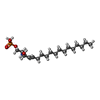 ChemComp-S1P: |
-Macromolecule #3: SULFATE ION
| Macromolecule | Name: SULFATE ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: SO4 |
|---|---|
| Molecular weight | Theoretical: 96.063 Da |
| Chemical component information |  ChemComp-SO4: |
-Macromolecule #4: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
| Macromolecule | Name: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine / type: ligand / ID: 4 / Number of copies: 2 / Formula: LBN |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-LBN: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)