[English] 日本語
 Yorodumi
Yorodumi- EMDB-23326: Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ATP... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23326 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ATP and 16x(Asp-Arg) | |||||||||
 Map data Map data | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ADPCP and 16x(Asp-Arg) map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationcyanophycin synthase (L-aspartate-adding) / cyanophycin synthase (L-arginine-adding) / cyanophycin synthetase activity (L-aspartate-adding) / cyanophycin synthetase activity (L-arginine-adding) / biosynthetic process / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
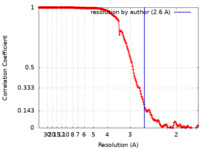
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Sharon I / Grogg M / Hilvert D / Schmeing TM | |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2021 Journal: Nat Chem Biol / Year: 2021Title: Structures and function of the amino acid polymerase cyanophycin synthetase. Authors: Itai Sharon / Asfarul S Haque / Marcel Grogg / Indrajit Lahiri / Dieter Seebach / Andres E Leschziner / Donald Hilvert / T Martin Schmeing /    Abstract: Cyanophycin is a natural biopolymer produced by a wide range of bacteria, consisting of a chain of poly-L-Asp residues with L-Arg residues attached to the β-carboxylate sidechains by isopeptide ...Cyanophycin is a natural biopolymer produced by a wide range of bacteria, consisting of a chain of poly-L-Asp residues with L-Arg residues attached to the β-carboxylate sidechains by isopeptide bonds. Cyanophycin is synthesized from ATP, aspartic acid and arginine by a homooligomeric enzyme called cyanophycin synthetase (CphA1). CphA1 has domains that are homologous to glutathione synthetases and muramyl ligases, but no other structural information has been available. Here, we present cryo-electron microscopy and X-ray crystallography structures of cyanophycin synthetases from three different bacteria, including cocomplex structures of CphA1 with ATP and cyanophycin polymer analogs at 2.6 Å resolution. These structures reveal two distinct tetrameric architectures, show the configuration of active sites and polymer-binding regions, indicate dynamic conformational changes and afford insight into catalytic mechanism. Accompanying biochemical interrogation of substrate binding sites, catalytic centers and oligomerization interfaces combine with the structures to provide a holistic understanding of cyanophycin biosynthesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23326.map.gz emd_23326.map.gz | 483.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23326-v30.xml emd-23326-v30.xml emd-23326.xml emd-23326.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23326_fsc.xml emd_23326_fsc.xml | 17.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_23326.png emd_23326.png | 211.8 KB | ||
| Masks |  emd_23326_msk_1.map emd_23326_msk_1.map | 512 MB |  Mask map Mask map | |
| Others |  emd_23326_half_map_1.map.gz emd_23326_half_map_1.map.gz emd_23326_half_map_2.map.gz emd_23326_half_map_2.map.gz | 474.5 MB 474.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23326 http://ftp.pdbj.org/pub/emdb/structures/EMD-23326 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23326 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23326 | HTTPS FTP |
-Validation report
| Summary document |  emd_23326_validation.pdf.gz emd_23326_validation.pdf.gz | 496.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23326_full_validation.pdf.gz emd_23326_full_validation.pdf.gz | 496 KB | Display | |
| Data in XML |  emd_23326_validation.xml.gz emd_23326_validation.xml.gz | 26.6 KB | Display | |
| Data in CIF |  emd_23326_validation.cif.gz emd_23326_validation.cif.gz | 34.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23326 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23326 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23326 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23326 | HTTPS FTP |
-Related structure data
| Related structure data |  7txuM  7lg5C  7lgjC  7lgmC  7lgnC  7lgqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23326.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23326.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ADPCP and 16x(Asp-Arg) map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.855 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
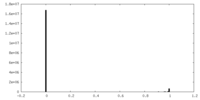
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23326_msk_1.map emd_23326_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
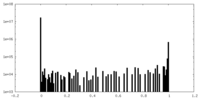
| Density Histograms |
-Half map: Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with...
| File | emd_23326_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ADPCP and 16x(Asp-Arg) half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
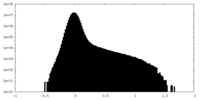
| Density Histograms |
-Half map: Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with...
| File | emd_23326_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ADPCP and 16x(Asp-Arg) half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
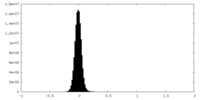
| Density Histograms |
- Sample components
Sample components
-Entire : Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ATP...
| Entire | Name: Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ATP and 16x(Asp-Arg) |
|---|---|
| Components |
|
-Supramolecule #1: Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ATP...
| Supramolecule | Name: Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ATP and 16x(Asp-Arg) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #1: Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ATP...
| Macromolecule | Name: Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ATP and 16x(Asp-Arg) type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MKILKTLTLR GPNYWSIRRK KLIVMRLDLE DLAERPSNSI PGFYEGLIKV LPSLVEHFCS PGYQGGFLER VKEGTYMGHI VEHVALELQE LVGMTAGFGR TRETSTPGV YNVVYEYVDE QAGRYAGRAA VRLCRSLVDT GDYPRLELEK DLEDLRDLGA NSALGPSTET ...String: MKILKTLTLR GPNYWSIRRK KLIVMRLDLE DLAERPSNSI PGFYEGLIKV LPSLVEHFCS PGYQGGFLER VKEGTYMGHI VEHVALELQE LVGMTAGFGR TRETSTPGV YNVVYEYVDE QAGRYAGRAA VRLCRSLVDT GDYPRLELEK DLEDLRDLGA NSALGPSTET IVTEAEARKI PWMLLSARAM VQLGYGVYQQ R IQATLSSH SGILGVELAC DKEGTKTILQ DAGIPVPRGT TIQYFDDLEE AINDVGGYPV VIKPLDGNHG RGITINVRHW QEAIAAYDLA AEESKSRAII VE RYYEGSD HRVLVVNGKL VAVAERIPAH VTGDGSSTIS ELIEKTNQDP NRGDGHDNIL TKIVVNKTAI DVMERQGYNL DSVLPKDEVV YLRATANLST GGI AIDRTD DIHPENIWLM ERVAKVIGLD IAGIDVVTSD ISKPLRETNG VIVEVNAAPG FRMHVAPSQG LPRNVAAPVL DMLFPPGTPS RIPILAVTGT NGKT TTTRL LAHIYRQTGK TVGYTSTDAI YINEYCVEKG DNTGPQSAGV ILRDPTVEVA VLETARGGIL RAGLAFDSCD VGVVLNVAAD HLGLGDIDTI EQMAK VKSV IAEVVDPSGY AVLNADDPLV AAMADKVKAK VAYFSMNPDN PIIQAHVRRN GIAAVYESGY LSILEGSWTL RVEQAKLIPM TMGGMAPFMI ANALAA CLA AFVNGLDVEV IRQGVRTFTT SAEQTPGRMN LFNLGQHHAL VDYAHNPAGY RAVGDFVKNW QGQRFGVVGG PGDRRDSDLI ELGQIAAQVF DRIIVKE DD DKRGRSEGET ADLIVKGILQ ENPGASYEVI LDETIALNKA LDQVEEKGLV VVFPESVTRA IDLIKVRNPI GENLYFQ |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)