+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | native AMPA receptor | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Yu J / Rao P / Gouaux E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition. Authors: Jie Yu / Prashant Rao / Sarah Clark / Jaba Mitra / Taekjip Ha / Eric Gouaux /  Abstract: AMPA-selective glutamate receptors mediate the transduction of signals between the neuronal circuits of the hippocampus. The trafficking, localization, kinetics and pharmacology of AMPA receptors are ...AMPA-selective glutamate receptors mediate the transduction of signals between the neuronal circuits of the hippocampus. The trafficking, localization, kinetics and pharmacology of AMPA receptors are tuned by an ensemble of auxiliary protein subunits, which are integral membrane proteins that associate with the receptor to yield bona fide receptor signalling complexes. Thus far, extensive studies of recombinant AMPA receptor-auxiliary subunit complexes using engineered protein constructs have not been able to faithfully elucidate the molecular architecture of hippocampal AMPA receptor complexes. Here we obtain mouse hippocampal, calcium-impermeable AMPA receptor complexes using immunoaffinity purification and use single-molecule fluorescence and cryo-electron microscopy experiments to elucidate three major AMPA receptor-auxiliary subunit complexes. The GluA1-GluA2, GluA1-GluA2-GluA3 and GluA2-GluA3 receptors are the predominant assemblies, with the auxiliary subunits TARP-γ8 and CNIH2-SynDIG4 non-stochastically positioned at the B'/D' and A'/C' positions, respectively. We further demonstrate how the receptor-TARP-γ8 stoichiometry explains the mechanism of and submaximal inhibition by a clinically relevant, brain-region-specific allosteric inhibitor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23285.map.gz emd_23285.map.gz | 483.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23285-v30.xml emd-23285-v30.xml emd-23285.xml emd-23285.xml | 30.2 KB 30.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23285.png emd_23285.png | 109.4 KB | ||
| Others |  emd_23285_additional_1.map.gz emd_23285_additional_1.map.gz emd_23285_additional_2.map.gz emd_23285_additional_2.map.gz emd_23285_additional_3.map.gz emd_23285_additional_3.map.gz | 199.9 MB 215.9 MB 483.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23285 http://ftp.pdbj.org/pub/emdb/structures/EMD-23285 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23285 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23285 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23285.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23285.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.00687 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_23285_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #3
| File | emd_23285_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #2
| File | emd_23285_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : GluA1/A2/A3-asymmetric-conformation1 recognized by antibody fragments
+Supramolecule #1: GluA1/A2/A3-asymmetric-conformation1 recognized by antibody fragments
+Supramolecule #2: GluA1/A2/A3-asymmetric-conformation1
+Supramolecule #3: 15F1 Fab
+Supramolecule #4: other antibody fragments
+Macromolecule #1: Glutamate receptor 1
+Macromolecule #2: Glutamate receptor
+Macromolecule #3: Glutamate receptor 3
+Macromolecule #4: Protein cornichon homolog 2
+Macromolecule #5: Voltage-dependent calcium channel gamma-8 subunit
+Macromolecule #6: 11B8 scFv
+Macromolecule #7: 15F1 Fab light chain
+Macromolecule #8: 15F1 Fab heavy chain
+Macromolecule #9: 5B2 Fab
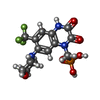
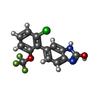
+Macromolecule #12: {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquino...
+Macromolecule #13: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #14: beta-D-mannopyranose
+Macromolecule #15: 6-[2-chloro-6-(trifluoromethoxy)phenyl]-1H-benzimidazol-2-ol
+Macromolecule #16: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 392000 |
|---|---|
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)