[English] 日本語
 Yorodumi
Yorodumi- EMDB-23007: Structure of SARS-CoV-2 backtracked complex bound to nsp13 helica... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23007 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of SARS-CoV-2 backtracked complex bound to nsp13 helicase - nsp13(1)-BTC | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA-dependent RNA polymerase / viral replication-transcription complex / transcription / viral proteins / TRANSFERASE-HYDROLASE-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / 5'-3' RNA helicase activity / Lyases; Phosphorus-oxygen lyases / mRNA guanylyltransferase activity / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / TRAF3-dependent IRF activation pathway / ISG15-specific peptidase activity ...protein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / 5'-3' RNA helicase activity / Lyases; Phosphorus-oxygen lyases / mRNA guanylyltransferase activity / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / TRAF3-dependent IRF activation pathway / ISG15-specific peptidase activity / Transcription of SARS-CoV-2 sgRNAs / snRNP Assembly / Translation of Replicase and Assembly of the Replication Transcription Complex / Replication of the SARS-CoV-2 genome / Hydrolases; Acting on ester bonds; Exoribonucleases producing 5'-phosphomonoesters / double membrane vesicle viral factory outer membrane / SARS coronavirus main proteinase / host cell endoplasmic reticulum-Golgi intermediate compartment / host cell endosome / 3'-5'-RNA exonuclease activity / 5'-3' DNA helicase activity / symbiont-mediated degradation of host mRNA / mRNA guanylyltransferase / symbiont-mediated suppression of host ISG15-protein conjugation / symbiont-mediated suppression of host toll-like receptor signaling pathway / G-quadruplex RNA binding / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / omega peptidase activity / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / SARS-CoV-2 modulates host translation machinery / symbiont-mediated suppression of host NF-kappaB cascade / host cell Golgi apparatus / DNA helicase / symbiont-mediated perturbation of host ubiquitin-like protein modification / methyltransferase cap1 activity / ubiquitinyl hydrolase 1 / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / cysteine-type deubiquitinase activity / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / lyase activity / single-stranded RNA binding / viral protein processing / host cell perinuclear region of cytoplasm / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / copper ion binding / symbiont-mediated suppression of host gene expression / symbiont-mediated activation of host autophagy / viral translational frameshifting / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / lipid binding / host cell nucleus / SARS-CoV-2 activates/modulates innate and adaptive immune responses / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
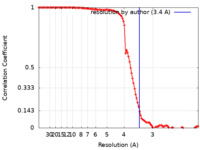
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Chen J / Malone B | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex. Authors: Brandon Malone / James Chen / Qi Wang / Eliza Llewellyn / Young Joo Choi / Paul Dominic B Olinares / Xinyun Cao / Carolina Hernandez / Edward T Eng / Brian T Chait / David E Shaw / Robert ...Authors: Brandon Malone / James Chen / Qi Wang / Eliza Llewellyn / Young Joo Choi / Paul Dominic B Olinares / Xinyun Cao / Carolina Hernandez / Edward T Eng / Brian T Chait / David E Shaw / Robert Landick / Seth A Darst / Elizabeth A Campbell /  Abstract: Backtracking, the reverse motion of the transcriptase enzyme on the nucleic acid template, is a universal regulatory feature of transcription in cellular organisms but its role in viruses is not ...Backtracking, the reverse motion of the transcriptase enzyme on the nucleic acid template, is a universal regulatory feature of transcription in cellular organisms but its role in viruses is not established. Here we present evidence that backtracking extends into the viral realm, where backtracking by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA-dependent RNA polymerase (RdRp) may aid viral transcription and replication. Structures of SARS-CoV-2 RdRp bound to the essential nsp13 helicase and RNA suggested the helicase facilitates backtracking. We use cryo-electron microscopy, RNA-protein cross-linking, and unbiased molecular dynamics simulations to characterize SARS-CoV-2 RdRp backtracking. The results establish that the single-stranded 3' segment of the product RNA generated by backtracking extrudes through the RdRp nucleoside triphosphate (NTP) entry tunnel, that a mismatched nucleotide at the product RNA 3' end frays and enters the NTP entry tunnel to initiate backtracking, and that nsp13 stimulates RdRp backtracking. Backtracking may aid proofreading, a crucial process for SARS-CoV-2 resistance against antivirals. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23007.map.gz emd_23007.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23007-v30.xml emd-23007-v30.xml emd-23007.xml emd-23007.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23007_fsc.xml emd_23007_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_23007.png emd_23007.png | 50.5 KB | ||
| Filedesc metadata |  emd-23007.cif.gz emd-23007.cif.gz | 7.8 KB | ||
| Others |  emd_23007_additional_1.map.gz emd_23007_additional_1.map.gz | 105.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23007 http://ftp.pdbj.org/pub/emdb/structures/EMD-23007 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23007 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23007 | HTTPS FTP |
-Related structure data
| Related structure data |  7krnMC  7kroC  7krpC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23007.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23007.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_23007_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : SARS-CoV-2 backtracked complex bound to nsp13 helicase - nsp13(1)-BTC
+Supramolecule #1: SARS-CoV-2 backtracked complex bound to nsp13 helicase - nsp13(1)-BTC
+Macromolecule #1: RNA-directed RNA polymerase
+Macromolecule #2: Non-structural protein 8
+Macromolecule #3: Non-structural protein 7
+Macromolecule #4: Helicase
+Macromolecule #5: RNA (37-MER)
+Macromolecule #6: RNA (43-MER)
+Macromolecule #7: ZINC ION
+Macromolecule #8: MAGNESIUM ION
+Macromolecule #9: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #10: CHAPSO
+Macromolecule #11: ALUMINUM FLUORIDE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)