[English] 日本語
 Yorodumi
Yorodumi- EMDB-22985: Negative stain electron microscopy reconstruction of 2P SARS-CoV-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22985 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain electron microscopy reconstruction of 2P SARS-CoV-2 spike ectodomain in complex with Fabs DH1043, DH1047, and DH1050.1 | |||||||||
 Map data Map data | Final sharpened map of 2P SARS-CoV-2 spike ectodomain in complex with Fabs DH1043, DH1047, and DH1050.1 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
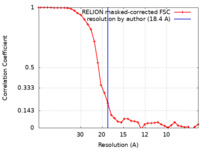
| Method | single particle reconstruction / negative staining / Resolution: 18.4 Å | |||||||||
 Authors Authors | Edwards RJ / Mansouri K | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2021 Journal: bioRxiv / Year: 2021Title: The functions of SARS-CoV-2 neutralizing and infection-enhancing antibodies in vitro and in mice and nonhuman primates. Authors: Dapeng Li / Robert J Edwards / Kartik Manne / David R Martinez / Alexandra Schäfer / S Munir Alam / Kevin Wiehe / Xiaozhi Lu / Robert Parks / Laura L Sutherland / Thomas H Oguin / Charlene ...Authors: Dapeng Li / Robert J Edwards / Kartik Manne / David R Martinez / Alexandra Schäfer / S Munir Alam / Kevin Wiehe / Xiaozhi Lu / Robert Parks / Laura L Sutherland / Thomas H Oguin / Charlene McDanal / Lautaro G Perez / Katayoun Mansouri / Sophie M C Gobeil / Katarzyna Janowska / Victoria Stalls / Megan Kopp / Fangping Cai / Esther Lee / Andrew Foulger / Giovanna E Hernandez / Aja Sanzone / Kedamawit Tilahun / Chuancang Jiang / Longping V Tse / Kevin W Bock / Mahnaz Minai / Bianca M Nagata / Kenneth Cronin / Victoria Gee-Lai / Margaret Deyton / Maggie Barr / Tarra Von Holle / Andrew N Macintyre / Erica Stover / Jared Feldman / Blake M Hauser / Timothy M Caradonna / Trevor D Scobey / Wes Rountree / Yunfei Wang / M Anthony Moody / Derek W Cain / C Todd DeMarco / ThomasN Denny / Christopher W Woods / Elizabeth W Petzold / Aaron G Schmidt / I-Ting Teng / Tongqing Zhou / Peter D Kwong / John R Mascola / Barney S Graham / Ian N Moore / Robert Seder / Hanne Andersen / Mark G Lewis / David C Montefiori / Gregory D Sempowski / Ralph S Baric / Priyamvada Acharya / Barton F Haynes / Kevin O Saunders Abstract: SARS-CoV-2 neutralizing antibodies (NAbs) protect against COVID-19. A concern regarding SARS-CoV-2 antibodies is whether they mediate disease enhancement. Here, we isolated NAbs against the receptor- ...SARS-CoV-2 neutralizing antibodies (NAbs) protect against COVID-19. A concern regarding SARS-CoV-2 antibodies is whether they mediate disease enhancement. Here, we isolated NAbs against the receptor-binding domain (RBD) and the N-terminal domain (NTD) of SARS-CoV-2 spike from individuals with acute or convalescent SARS-CoV-2 or a history of SARS-CoV-1 infection. Cryo-electron microscopy of RBD and NTD antibodies demonstrated function-specific modes of binding. Select RBD NAbs also demonstrated Fc receptor-γ (FcγR)-mediated enhancement of virus infection , while five non-neutralizing NTD antibodies mediated FcγR-independent infection enhancement. However, both types of infection-enhancing antibodies protected from SARS-CoV-2 replication in monkeys and mice. Nonetheless, three of 31 monkeys infused with enhancing antibodies had higher lung inflammation scores compared to controls. One monkey had alveolar edema and elevated bronchoalveolar lavage inflammatory cytokines. Thus, while antibody-enhanced infection does not necessarily herald enhanced infection , increased lung inflammation can occur in SARS-CoV-2 antibody-infused macaques. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22985.map.gz emd_22985.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22985-v30.xml emd-22985-v30.xml emd-22985.xml emd-22985.xml | 30 KB 30 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22985_fsc.xml emd_22985_fsc.xml | 3.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_22985.png emd_22985.png | 80.7 KB | ||
| Masks |  emd_22985_msk_1.map emd_22985_msk_1.map | 3.4 MB |  Mask map Mask map | |
| Others |  emd_22985_additional_1.map.gz emd_22985_additional_1.map.gz emd_22985_half_map_1.map.gz emd_22985_half_map_1.map.gz emd_22985_half_map_2.map.gz emd_22985_half_map_2.map.gz | 2.4 MB 2.4 MB 2.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22985 http://ftp.pdbj.org/pub/emdb/structures/EMD-22985 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22985 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22985 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22985.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22985.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final sharpened map of 2P SARS-CoV-2 spike ectodomain in complex with Fabs DH1043, DH1047, and DH1050.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.02 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22985_msk_1.map emd_22985_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened map of 2P SARS-CoV-2 spike ectodomain in...
| File | emd_22985_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map of 2P SARS-CoV-2 spike ectodomain in complex with Fabs DH1043, DH1047, and DH1050.1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1 of 2P SARS-CoV-2 spike ectodomain in...
| File | emd_22985_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 of 2P SARS-CoV-2 spike ectodomain in complex with Fabs DH1043, DH1047, and DH1050.1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2of 2P SARS-CoV-2 spike ectodomain in complex...
| File | emd_22985_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2of 2P SARS-CoV-2 spike ectodomain in complex with Fabs DH1043, DH1047, and DH1050.1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 2P SARS-CoV-2 spike ectodomain in complex with Fabs DH1043, DH104...
| Entire | Name: 2P SARS-CoV-2 spike ectodomain in complex with Fabs DH1043, DH1047, and DH1050.1 |
|---|---|
| Components |
|
-Supramolecule #1: 2P SARS-CoV-2 spike ectodomain in complex with Fabs DH1043, DH104...
| Supramolecule | Name: 2P SARS-CoV-2 spike ectodomain in complex with Fabs DH1043, DH1047, and DH1050.1 type: complex / ID: 1 / Parent: 0 |
|---|---|
| Molecular weight | Theoretical: 481.8 KDa |
-Supramolecule #2: 2P SARS-CoV-2 spike ectodomain
| Supramolecule | Name: 2P SARS-CoV-2 spike ectodomain / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: 293F / Recombinant plasmid: p-alpha-H Homo sapiens (human) / Recombinant cell: 293F / Recombinant plasmid: p-alpha-H |
-Supramolecule #3: Fab DH1043
| Supramolecule | Name: Fab DH1043 / type: complex / ID: 3 / Parent: 1 Details: Fab fragment generated by proteolytic cleavage of IgG antibody |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: Fab DH1047
| Supramolecule | Name: Fab DH1047 / type: complex / ID: 4 / Parent: 1 Details: Fab fragment generated by proteolytic cleavage of IgG antibody |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #5: Fab DH1050.1
| Supramolecule | Name: Fab DH1050.1 / type: complex / ID: 5 / Parent: 1 Details: Fab fragment generated by proteolytic cleavage of IgG antibody |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293i Homo sapiens (human) / Recombinant cell: HEK293i |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: Samples were diluted to 0.1 mg/mL in 20 mM HEPES buffer, pH 7.4, with 5% glycerol, 150 mM NaCl, and 7.5 mM glutaraldehyde. After 5 minute incubation at room temperature, sufficient 1 M Tris ...Details: Samples were diluted to 0.1 mg/mL in 20 mM HEPES buffer, pH 7.4, with 5% glycerol, 150 mM NaCl, and 7.5 mM glutaraldehyde. After 5 minute incubation at room temperature, sufficient 1 M Tris stock, pH 7.4, was added to a final concentration of 75 mM Tris to quench unreacted glutaraldehyde, and was then incubated 5 minutes. Sample was then applied to a carbon film over 400 mesh copper EM grids that had been glow-discharged, incubated 1 minute, and then stained with 2% uranyl formate. | ||||||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5.0 nm / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS EM420 |
|---|---|
| Image recording | Film or detector model: OTHER / Digitization - Dimensions - Width: 2048 pixel / Digitization - Dimensions - Height: 2048 pixel / Digitization - Sampling interval: 33.0 µm / Number grids imaged: 1 / Number real images: 520 / Average exposure time: 0.5 sec. / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 82000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera





































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)