[English] 日本語
 Yorodumi
Yorodumi- EMDB-22934: Negative stain electron microscopy reconstruction of rS2d-hexapro... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22934 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
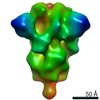
| Title | Negative stain electron microscopy reconstruction of rS2d-hexapro SARS-CoV-2 spike ectodomain | |||||||||
 Map data Map data | Unsharpened map of rS2d-hexapro SARS-CoV-2 spike ectodomain | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
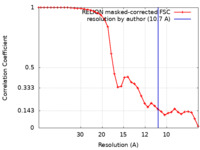
| Method | single particle reconstruction / negative staining / Resolution: 10.7 Å | |||||||||
 Authors Authors | Edwards RJ / Mansouri K | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2020 Journal: bioRxiv / Year: 2020Title: Cold sensitivity of the SARS-CoV-2 spike ectodomain. Authors: Robert J Edwards / Katayoun Mansouri / Victoria Stalls / Kartik Manne / Brian Watts / Rob Parks / Katarzyna Janowska / Sophie M C Gobeil / Megan Kopp / Dapeng Li / Xiaozhi Lu / Zekun Mu / ...Authors: Robert J Edwards / Katayoun Mansouri / Victoria Stalls / Kartik Manne / Brian Watts / Rob Parks / Katarzyna Janowska / Sophie M C Gobeil / Megan Kopp / Dapeng Li / Xiaozhi Lu / Zekun Mu / Margaret Deyton / Thomas H Oguin / Jordan Sprenz / Wilton Williams / Kevin Saunders / David Montefiori / Gregory D Sempowski / Rory Henderson / Munir Alam / Barton F Haynes / Priyamvada Acharya Abstract: The SARS-CoV-2 spike (S) protein, a primary target for COVID-19 vaccine development, presents its Receptor Binding Domain in two conformations: receptor-accessible "up" or receptor-inaccessible ...The SARS-CoV-2 spike (S) protein, a primary target for COVID-19 vaccine development, presents its Receptor Binding Domain in two conformations: receptor-accessible "up" or receptor-inaccessible "down" conformations. Here, we report that the commonly used stabilized S ectodomain construct "2P" is sensitive to cold temperature, and that this cold sensitivity is resolved in a "down" state stabilized spike. Our results will impact structural, functional and vaccine studies that use the SARS-CoV-2 S ectodomain. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22934.map.gz emd_22934.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22934-v30.xml emd-22934-v30.xml emd-22934.xml emd-22934.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22934_fsc.xml emd_22934_fsc.xml | 3.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_22934.png emd_22934.png | 47.8 KB | ||
| Masks |  emd_22934_msk_1.map emd_22934_msk_1.map | 3.4 MB |  Mask map Mask map | |
| Others |  emd_22934_additional_1.map.gz emd_22934_additional_1.map.gz emd_22934_half_map_1.map.gz emd_22934_half_map_1.map.gz emd_22934_half_map_2.map.gz emd_22934_half_map_2.map.gz | 2.4 MB 2.4 MB 2.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22934 http://ftp.pdbj.org/pub/emdb/structures/EMD-22934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22934 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22934.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22934.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map of rS2d-hexapro SARS-CoV-2 spike ectodomain | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.02 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22934_msk_1.map emd_22934_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened map of rS2d-hexapro SARS-CoV-2 spike ectodomain
| File | emd_22934_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map of rS2d-hexapro SARS-CoV-2 spike ectodomain | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2 of rS2d-hexapro SARS-CoV-2 spike ectodomain
| File | emd_22934_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 of rS2d-hexapro SARS-CoV-2 spike ectodomain | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1 of rS2d-hexapro SARS-CoV-2 spike ectodomain
| File | emd_22934_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 of rS2d-hexapro SARS-CoV-2 spike ectodomain | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 rS2d Down State Spike Protein Trimer
| Entire | Name: SARS-CoV-2 rS2d Down State Spike Protein Trimer |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 rS2d Down State Spike Protein Trimer
| Supramolecule | Name: SARS-CoV-2 rS2d Down State Spike Protein Trimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: 293F / Recombinant plasmid: p-alpha-H Homo sapiens (human) / Recombinant cell: 293F / Recombinant plasmid: p-alpha-H |
| Molecular weight | Theoretical: 481.8 KDa |
-Macromolecule #1: rS2d-hexapro SARS-CoV-2 spike ectodomain
| Macromolecule | Name: rS2d-hexapro SARS-CoV-2 spike ectodomain / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY VSQPFLMDLE GKQGNFKNLR EFVFKNIDGY FKIYSKHTPI NLVRDLPQGF SALEPLVDLP IGINITRFQT LLALHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPTESIVRFP NITNLCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVCPTKLNDL CFTNVYADSF VIRGDEVRQI APGQTGKIAD YNYKLPDDFT GCVIAWNSNN LDSKVGGNYN YLYRLFRKSN LKPFERDIST EIYQAGSTPC NGVEGFNCYF PLQSYGFQPT NGVGYQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLTGTG VLTESNKKFL PFQQFGRDIA DTTDAVRDPQ TLEILDITPC SFGGVSVITP GTNTSNQVAV LYQDVNCTEV PVAIHADQLT PTWRVYSTGS NVFQTRAGCL IGAEHVNNSY ECDIPIGAGI CASYQTQTNS PGSASSVASQ SIIAYTMSLG AENSVAYSNN SIAIPTNFTI SVTTEILPVS MTKTSVDCTM YICGDSTECS NLLLQYGSFC TQLNRALTGI AVEQDKNTQE VFAQVKQIYK TPPIKDFGGF NFSQILPDPS KPSKRSPIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFGA GPALQIPFPM QMAYRFNGIG VTQNVLYENQ KLIANQFNSA IGKIQDSLSS TPSALGKLQD VVNQNAQALN TLVKQLSSNF GAISSVLNDI LSRLCPPEAE VQIDRLITGR LQSLQTYVTQ QLIRAAEIRA SANLAATKMS ECVLGQSKRV DFCGKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVSGNCDVVI GIVNNTVYDP LQPELDSFKE ELDKYFKNHT SPDVDLGDIS GINASVVNIQ KEIDRLNEVA KNLNESLIDL QELGKYEQ |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: Samples were diluted to 0.1 mg/mL in 20 mM HEPES buffer, pH 7.4, with 5% glycerol, 150 mM NaCl, and 7.5 mM glutaraldehyde. After 5 minute incubation at room temperature, sufficient 1 M Tris ...Details: Samples were diluted to 0.1 mg/mL in 20 mM HEPES buffer, pH 7.4, with 5% glycerol, 150 mM NaCl, and 7.5 mM glutaraldehyde. After 5 minute incubation at room temperature, sufficient 1 M Tris stock, pH 7.4, was added to a final concentration of 75 mM Tris to quench unreacted glutaraldehyde, and was then incubated 5 minutes. Sample was then applied to a carbon film over 400 mesh copper EM grids that had been glow-discharged, incubated 1 minute, and then stained with 2% uranyl formate. | ||||||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.05 nm / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS EM420 |
|---|---|
| Image recording | Film or detector model: OTHER / Digitization - Dimensions - Width: 2048 pixel / Digitization - Dimensions - Height: 2048 pixel / Digitization - Sampling interval: 33.0 µm / Number grids imaged: 1 / Number real images: 145 / Average exposure time: 0.5 sec. / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 82000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)