+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22890 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Plasmodium falciparum RhopH complex in soluble form | |||||||||
 Map data Map data | Unsharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Plasmodium falciparum / malaria / RhopH complex / MEMBRANE PROTEIN | |||||||||
| Function / homology | : / RhopH3 C-terminal domain / rhoptry / symbiont-containing vacuole membrane / cytoplasmic vesicle / host cell cytoplasm / host cell plasma membrane / RhopH3 C-terminal domain-containing protein / High molecular weight rhoptry protein 2 Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Schureck MA / Darling JE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Malaria parasites use a soluble RhopH complex for erythrocyte invasion and an integral form for nutrient uptake. Authors: Marc A Schureck / Joseph E Darling / Alan Merk / Jinfeng Shao / Geervani Daggupati / Prakash Srinivasan / Paul Dominic B Olinares / Michael P Rout / Brian T Chait / Kurt Wollenberg / Sriram ...Authors: Marc A Schureck / Joseph E Darling / Alan Merk / Jinfeng Shao / Geervani Daggupati / Prakash Srinivasan / Paul Dominic B Olinares / Michael P Rout / Brian T Chait / Kurt Wollenberg / Sriram Subramaniam / Sanjay A Desai /   Abstract: Malaria parasites use the RhopH complex for erythrocyte invasion and channel-mediated nutrient uptake. As the member proteins are unique to Plasmodium spp., how they interact and traffic through ...Malaria parasites use the RhopH complex for erythrocyte invasion and channel-mediated nutrient uptake. As the member proteins are unique to Plasmodium spp., how they interact and traffic through subcellular sites to serve these essential functions is unknown. We show that RhopH is synthesized as a soluble complex of CLAG3, RhopH2, and RhopH3 with 1:1:1 stoichiometry. After transfer to a new host cell, the complex crosses a vacuolar membrane surrounding the intracellular parasite and becomes integral to the erythrocyte membrane through a PTEX translocon-dependent process. We present a 2.9 Å single-particle cryo-electron microscopy structure of the trafficking complex, revealing that CLAG3 interacts with the other subunits over large surface areas. This soluble complex is tightly assembled with extensive disulfide bonding and predicted transmembrane helices shielded. We propose a large protein complex stabilized for trafficking but poised for host membrane insertion through large-scale rearrangements, paralleling smaller two-state pore-forming proteins in other organisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22890.map.gz emd_22890.map.gz | 194 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22890-v30.xml emd-22890-v30.xml emd-22890.xml emd-22890.xml | 24.7 KB 24.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22890_fsc.xml emd_22890_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_22890.png emd_22890.png | 232.7 KB | ||
| Filedesc metadata |  emd-22890.cif.gz emd-22890.cif.gz | 8.1 KB | ||
| Others |  emd_22890_additional_1.map.gz emd_22890_additional_1.map.gz emd_22890_additional_2.map.gz emd_22890_additional_2.map.gz emd_22890_half_map_1.map.gz emd_22890_half_map_1.map.gz emd_22890_half_map_2.map.gz emd_22890_half_map_2.map.gz | 228.7 MB 146.9 MB 194.3 MB 194.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22890 http://ftp.pdbj.org/pub/emdb/structures/EMD-22890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22890 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22890 | HTTPS FTP |
-Related structure data
| Related structure data |  7kiyMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22890.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22890.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Sharpened map
| File | emd_22890_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
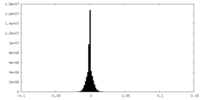
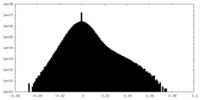
| Density Histograms |
-Additional map: Locally sharpened map
| File | emd_22890_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
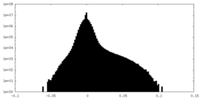
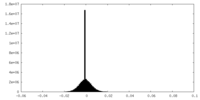
| Density Histograms |
-Half map: Half map 1
| File | emd_22890_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
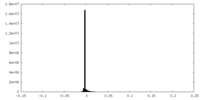
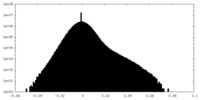
| Density Histograms |
-Half map: Half map 2
| File | emd_22890_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
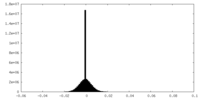
| Density Histograms |
- Sample components
Sample components
-Entire : RhopH complex
| Entire | Name: RhopH complex |
|---|---|
| Components |
|
-Supramolecule #1: RhopH complex
| Supramolecule | Name: RhopH complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 434 KDa |
-Macromolecule #1: Cytoadherence linked asexual protein 3
| Macromolecule | Name: Cytoadherence linked asexual protein 3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 177.069516 KDa |
| Sequence | String: MVSFFKTPII IFFFLLCLNE KVLCSINENE NLGENKNENA NVNTPENLNK LLNEYDNIEQ LKSMIGNDEL HKNLTILEKL ILESLEKDK LKYPLLKQGT EQLIDISKFN KKNITDADDE TYIIPTVQSS FHDIVKYEHL IKEQSIEIYN SDISDKIKKK I FIVRTLKT ...String: MVSFFKTPII IFFFLLCLNE KVLCSINENE NLGENKNENA NVNTPENLNK LLNEYDNIEQ LKSMIGNDEL HKNLTILEKL ILESLEKDK LKYPLLKQGT EQLIDISKFN KKNITDADDE TYIIPTVQSS FHDIVKYEHL IKEQSIEIYN SDISDKIKKK I FIVRTLKT IKLMLIPLNS YKQNNDLKSA LEELNNVFTN KEAQKESSPI GDHGTFFRKL LTHVRTIKEN EDIENKGETL IL GDNKIDV MNSNDFFFTT NSNVKFMENL DDITNQYGLG LINHLGPHLI ALGHFTVLKL ALKNYKNYFE AKSIKFFSWQ KIL EFSMSD RFKVLDMMCD HESVYYSEKK RRKTYLKVDR SNTSMECNIL EYLLHYFNKY QLEIIKTTQD TDFDLHGMME HKYI KDYFF SFMCNDPKEC IIYHTNQFKK EANEENTFPE QEEPNRQISA FNLYLNYYYF MKRYSSYGVK KTLYVHLLNL TGLLN YDTR SYVTSLYLPG YYNAVEMSFT EEKEFSKLFE SLIQCIEKCH SDQARQISKD SNLLNDITKC DLCKGAFLYS NMKFDE VPS MLQKFYLYLT KGLKIQKVSS LIKTLDIYQD YSNFLSHDIN WYTFLFLFRL TSFKEISKKN VAEAMYLNIK DEDTFNK TI VTNYWYPSPI KKYYTLYVRK HIPNNLVDEL EKLMKSGTLE KMKKSLTFLV HVNSFLQLDF FHQLNEPPLG LPRSYPLS L VLEHKFKEWM DSSPAGFYFS NYQNPYVRKD LHDKVLSQKF EPPKMNQWNK VLKSLIECAY DMYFEQRHVK NLYKYHNIY NINNKLMLMR DSIDLYKTHF DDVLFFADIF NMRKYMTATP VYKKVKDRVY HTLHSITGNS VNFYKYGIIY GFKVNKEILK EVVDELYSI YNFNTDIFTD TSFLQTVYLL FRRIEETYRT QRRDDKISVN NVFFMNVANN YSKLNKEERE IEIHNSMASR Y YAKTMFAA FQMLFSTMLS NNVDNLDKAY GLSENIQVAT STSAFLTFAY VYNGSIMDSV TNSLLPPYAK KPITQLKYGK TF VFSNYFM LASKMYDMLN YKNLSLLCEY QAVASANFYS AKKVGQFIGR KFLPITTYFL VMRISWTHAF TTGQHLIAAF DPL NTNTSP KPNGGSGIYK SPESFFFTHA LAAEASKYLF FYFFTNLYLD AYKSFPGGFG PAIKEQTQHV QEQTYERKPS VHSF NRNFF MELANGFMYA FCFFAISQMY AYFENINFYI TSNFRFLDRY YGVFNKYFIN YARIKLKEIT SDLLIKYERE AYLSM KKYG YLGEVIAARL SPKDKIMNYV HETNDDVMSN LRRYDMENAF KNKMSTYVDD FAFFDDCGKN EQFLNERCDY CPVIEE VEE TELFTTTGDK NTNKTTEIKK QTSTYIDTEK MNEADSADSD DEKDSDTPDN ELMIARFHGA GHHHHHHHHH HDYKDDD DK GLVPRGSAAA AYPYDVPDYA SAWSHPQFEK GGGSGGGSGG SAWSHPQFEK GPDRKAAVSH WQQ |
-Macromolecule #2: High molecular weight rhoptry protein-2
| Macromolecule | Name: High molecular weight rhoptry protein-2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 162.877406 KDa |
| Sequence | String: MIKVTIFLLL SIFSFNLYGL ELNEKVSIKY GAEQGVGSAD SNTKLCSDIL KYLYMDEYLS EGDKATFEKK CHNVIGNIRN TFSNKNTIK EGNEFLMSIL HMKSLYGNNN NNNAGSESDV TLKSLYLSLK GSQNTEGESE VPSDDEINKT IMNFVKFNKY L LDNSNDIK ...String: MIKVTIFLLL SIFSFNLYGL ELNEKVSIKY GAEQGVGSAD SNTKLCSDIL KYLYMDEYLS EGDKATFEKK CHNVIGNIRN TFSNKNTIK EGNEFLMSIL HMKSLYGNNN NNNAGSESDV TLKSLYLSLK GSQNTEGESE VPSDDEINKT IMNFVKFNKY L LDNSNDIK KVHDFLVLTS QSNENLLPNK EKLFEQIVDQ IKYFDEYFFA SGGKIKVKKG YLKYNFLDIY KQPVCSAYLH LC SRYYESV SIYIRLKKVF NGIPAFLDKN CRKVKGEEFK KLMDMELKHN HIVERFDKYI ISDDLYYVNM KVFDLKNVDK IQV SKIDDI NNLNIYEHKE TMHLSAKNLS RYIDIKKELN DEKAYKQLMS AIRKYVTTLT KADSDITYFV KQLDDEEIER FLID LNFFL YNGFLRITED KHLINADDVS PSYINLYRSN NIVALYILKT QYEENKLSEY RAHKFYRRKR VSNITNDMIK KDFTQ TNAL TNLPNLDNKK TTEYYLKEYE NFVENFQPDL HDIMKLQLFF TMAFKDCNVN QNFTETSKKL WFDLLYAYDK FGWFYI HPN EVINSINKTD FVRHVLVSRN FLLKNNDQLT FLETQVAKIV EIINLSLEVD KSPDSLDFSI PMNFFNHKNG YHVMNDD KL KLLTSYEYID SIANNYFFLS EYKNDVFRTG NNFKLYFNLP NIYSLAYQLF NELAININVI TNVPLKKYLK YNASYAYF T LMNMIGKNHD IYSKGSRFVY ASYILGLVFF IESHIDIARL KPKDFFFMKQ SLPIIDHVYH KDLKTLKKNC TLLTDFMKI NKNSQNYSLT HTEEMIKILG LLTVTLWAKE GKKSVYYDDD VSLYRKLMVS CVFNGGETIQ EKLANNIEKS CDISQYGIKS KNLKDMIDI NLSIHKWNPA EIEKLAYSFV LSCKMQKLMY KPMNVEKLPL EDYYKLPLAP DMVKTYHCYK LGKQAAKLLE S IILKKKFV RFRVTDAIDV YDFFYIKKVL SSHIKKEYNE FLQDKRAFEK KELETILNNS PFSEEQTMKL INSYECHWFT SY ENFRILW MHASSNLGTG TYLKNFFSEL WQNIRFLFKS KLKIRDMEYF SGDISQMNLL DYYSPMVHSE SHCQEKMQVL FIT LRDSKE ENRSEIAQKV KSAYYQCKLD YYKNHHSDFI HRIHPNDFLN NKVYVLKQPY YLMSNVPLNN PKKVSRLFVT EGTL EYLLL DKINIPECFG PCTKLHFNKV VIKESKQRIY DMTINNALVP EIQPYNRRKY MTIYINEAYI KNIVSDALTS EEIKR HDIQ KGNIKICMGK STYLTEPILT EEHFNLTHKP VYDFSSVKHN LKVFHMKNEH LVSEDPNDDC FINYPLATIN LDISDP YKE ISEDLIKNLY ILKSS UniProtKB: High molecular weight rhoptry protein 2 |
-Macromolecule #3: High molecular weight rhoptry protein 3
| Macromolecule | Name: High molecular weight rhoptry protein 3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 104.981406 KDa |
| Sequence | String: MRSKHLVTLF IITFLSFSTV KVWGKDVFAG FVTKKLKTLL DCNFALYYNF KGNGPDAGSF LDFVDEPEQF YWFVEHFLSV KFRVPKHLK DKNIHNFTPC LNRSWVSEFL KEYEEPFVNP VMKFLDKEQR LFFTYNFGDV EPQGKYTYFP VKEFHKYCIL P PLIKTNIK ...String: MRSKHLVTLF IITFLSFSTV KVWGKDVFAG FVTKKLKTLL DCNFALYYNF KGNGPDAGSF LDFVDEPEQF YWFVEHFLSV KFRVPKHLK DKNIHNFTPC LNRSWVSEFL KEYEEPFVNP VMKFLDKEQR LFFTYNFGDV EPQGKYTYFP VKEFHKYCIL P PLIKTNIK DGESGEFLKY QLNKEEYKVF LSSVGSQMTA IKNLYSTVED EQRKQLLKVI IENESTNDIS VQCPTYNIKL HY TKECANS NNILKCIDEF LRKTCEKKTE SKHPSADLCE HLQFLFESLK NPYLDNFKKF MTNSDFTLIK PQSVWNVPIF DIY KPKNYL DSVQNLDTEC FKKLNSKNLI FLSFHDDIPN NPYYNVELQE IVKLSTYTYS IFDKLYNFFF VFKKSGAPIS PVSV KELSH NITDFSFKED NSEIQCQNVR KSLDLEVDVE TMKGIAAEKL CKIIEKFILT KDDASKPEKS DIHRGFRILC ILIST HVEA YNIVRQLLNM ESMISLTRYT SLYIHKFFKS VTLLKGNFLY KNNKAIRYSR ACSKASLHVP SVLYRRNIYI PETFLS LYL GLSNLVSSNP SSPFFEYAII EFLVTYYNKG SEKFVLYFIS IISVLYINEY YYEQLSCFYP KEFELIKSRM IHPNIVD RI LKGIDNLMKS TRYDKMRTMY LDFESSDIFS REKVFTALYN FDSFIKTNEQ LKKKNLEEIS EIPVQLETSN DGIGYRKQ D VLYETDKPQT MDEASYEETV DEDAHHVNEK QHSAHFLDAI AEKDILEEKT KDQDLEIELY KYMGPLKEQS KSTSAASTS DELAGSEGPS TESTSTGNQG EDKTTDNTYK EMEELEEAEG TSNLKKGLEF YKSSLKLDQL DKEKPKKKKS KRKKKRDSSS DRILLEESK TFTSENEL UniProtKB: RhopH3 C-terminal domain-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-58 / Number grids imaged: 1 / Number real images: 1310 / Average exposure time: 0.4 sec. / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 97.81 |
|---|---|
| Output model |  PDB-7kiy: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)