[English] 日本語
 Yorodumi
Yorodumi- EMDB-22830: Murine core lysosomal multienzyme complex (LMC) composed of acid ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22830 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Murine core lysosomal multienzyme complex (LMC) composed of acid beta-galactosidase (GLB1) and protective protein cathepsin A (PPCA, CTSA) | ||||||||||||
 Map data Map data | unsharpened | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | glycosidase / protease / lysosome / HYDROLASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationKeratan sulfate degradation / HS-GAG degradation / keratan sulfate proteoglycan catabolic process / response to cortisone / Sialic acid metabolism / response to Thyroglobulin triiodothyronine / carboxypeptidase C / Glycosphingolipid catabolism / galactose catabolic process / serine-type carboxypeptidase activity ...Keratan sulfate degradation / HS-GAG degradation / keratan sulfate proteoglycan catabolic process / response to cortisone / Sialic acid metabolism / response to Thyroglobulin triiodothyronine / carboxypeptidase C / Glycosphingolipid catabolism / galactose catabolic process / serine-type carboxypeptidase activity / galactoside binding / glycoprotein catabolic process / beta-galactosidase / MHC class II antigen presentation / ganglioside catabolic process / beta-galactosidase activity / negative regulation of chaperone-mediated autophagy / Neutrophil degranulation / regulation of protein stability / lysosome / hydrolase activity / Golgi apparatus / protein homodimerization activity / mitochondrion / proteolysis / extracellular space / membrane / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.59 Å | ||||||||||||
 Authors Authors | Gorelik A / Illes K / Hasan SMN / Nagar B / Mazhab-Jafari MT | ||||||||||||
| Funding support |  Canada, 3 items Canada, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Structure of the murine lysosomal multienzyme complex core. Authors: Alexei Gorelik / Katalin Illes / S M Naimul Hasan / Bhushan Nagar / Mohammad T Mazhab-Jafari /  Abstract: The enzymes β-galactosidase (GLB1) and neuraminidase 1 (NEU1; sialidase 1) participate in the degradation of glycoproteins and glycolipids in the lysosome. To remain active and stable, they ...The enzymes β-galactosidase (GLB1) and neuraminidase 1 (NEU1; sialidase 1) participate in the degradation of glycoproteins and glycolipids in the lysosome. To remain active and stable, they associate with PPCA [protective protein cathepsin A (CTSA)] into a high-molecular weight lysosomal multienzyme complex (LMC), of which several forms exist. Genetic defects in these three proteins cause the lysosomal storage diseases GM1-gangliosidosis/mucopolysaccharidosis IV type B, sialidosis, and galactosialidosis, respectively. To better understand the interactions between these enzymes, we determined the three-dimensional structure of the murine LMC core. This 0.8-MDa complex is composed of three GLB1 dimers and three CTSA dimers, adopting a triangular architecture maintained through six copies of a unique GLB1-CTSA polar interface. Mutations in this contact surface that occur in GM1-gangliosidosis prevent formation of the LMC in vitro. These findings may facilitate development of therapies for lysosomal storage disorders. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22830.map.gz emd_22830.map.gz | 61.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22830-v30.xml emd-22830-v30.xml emd-22830.xml emd-22830.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22830.png emd_22830.png | 228.7 KB | ||
| Filedesc metadata |  emd-22830.cif.gz emd-22830.cif.gz | 7 KB | ||
| Others |  emd_22830_additional_1.map.gz emd_22830_additional_1.map.gz emd_22830_additional_2.map.gz emd_22830_additional_2.map.gz emd_22830_half_map_1.map.gz emd_22830_half_map_1.map.gz emd_22830_half_map_2.map.gz emd_22830_half_map_2.map.gz | 22.9 MB 113.7 MB 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22830 http://ftp.pdbj.org/pub/emdb/structures/EMD-22830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22830 | HTTPS FTP |
-Validation report
| Summary document |  emd_22830_validation.pdf.gz emd_22830_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22830_full_validation.pdf.gz emd_22830_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_22830_validation.xml.gz emd_22830_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_22830_validation.cif.gz emd_22830_validation.cif.gz | 16.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22830 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22830 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22830 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22830 | HTTPS FTP |
-Related structure data
| Related structure data |  7kdvMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22830.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22830.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
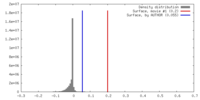
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: density-modified with Phenix ResolveCryoEM
| File | emd_22830_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | density-modified with Phenix ResolveCryoEM | ||||||||||||
| Projections & Slices |
| ||||||||||||
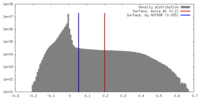
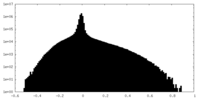
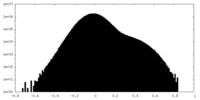
| Density Histograms |
-Additional map: sharpened with Phenix Autosharpen
| File | emd_22830_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened with Phenix Autosharpen | ||||||||||||
| Projections & Slices |
| ||||||||||||
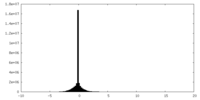
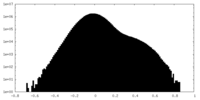
| Density Histograms |
-Half map: half-map A
| File | emd_22830_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
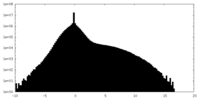
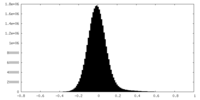
| Density Histograms |
-Half map: half-map B
| File | emd_22830_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
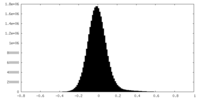
| Density Histograms |
- Sample components
Sample components
-Entire : core lysosomal multienzyme complex composed of CTSA and GLB1
| Entire | Name: core lysosomal multienzyme complex composed of CTSA and GLB1 |
|---|---|
| Components |
|
-Supramolecule #1: core lysosomal multienzyme complex composed of CTSA and GLB1
| Supramolecule | Name: core lysosomal multienzyme complex composed of CTSA and GLB1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Beta-galactosidase
| Macromolecule | Name: Beta-galactosidase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: beta-galactosidase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 71.45018 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DRHHHHHHGS VTQRTFKLDY SRDRFLKDGQ PFRYISGSIH YFRIPRFYWE DRLLKMKMAG LNAIQMYVPW NFHEPQPGQY EFSGDRDVE HFIQLAHELG LLVILRPGPY ICAEWDMGGL PAWLLEKQSI VLRSSDPDYL VAVDKWLAVL LPKMKPLLYQ N GGPIITVQ ...String: DRHHHHHHGS VTQRTFKLDY SRDRFLKDGQ PFRYISGSIH YFRIPRFYWE DRLLKMKMAG LNAIQMYVPW NFHEPQPGQY EFSGDRDVE HFIQLAHELG LLVILRPGPY ICAEWDMGGL PAWLLEKQSI VLRSSDPDYL VAVDKWLAVL LPKMKPLLYQ N GGPIITVQ VENEYGSYFA CDYDYLRFLV HRFRYHLGND VILFTTDGAS EKMLKCGTLQ DLYATVDFGT GNNITQAFLV QR KFEPKGP LINSEFYTGW LDHWGKPHST VKTKTLATSL YNLLARGANV NLYMFIGGTN FAYWNGANTP YEPQPTSYDY DAP LSEAGD LTKKYFALRE VIQMFKEVPE GPIPPSTPKF AYGKVALRKF KTVAEALGIL CPNGPVKSLY PLTFTQVKQY FGYV LYRTT LPQDCSNPKP IFSSPFNGVR DRAYVSVDGV PQGILDRNLM TALNIQGKAG ATLDILVENM GRVNYGRFIN DFKGL ISNM TINSTVLTNW TVFPLDTEAM VRNHLWGREA SDGGHLDGRS TSNSSDLILP TFYVGNFSIP SGIPDLPQDT FIQFPG WSK GQVWINGFNL GRYWPTMGPQ KTLFVPRNIL TTSAPNNITV LELEFAPCSE GTPELCTVEF VDTPVIS UniProtKB: Beta-galactosidase |
-Macromolecule #2: Lysosomal protective protein
| Macromolecule | Name: Lysosomal protective protein / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO / EC number: carboxypeptidase C |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.657141 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DRHHHHHHGS APDQDEIDAL PGLAKQPSFR QYSGYLRASD SKHFHYWFVE SQNDPKNSPV VLWLNGGPGC SSLDGLLTEH GPFLIQPDG VTLEYNPYAW NLIANVLYIE SPAGVGFSYS DDKMYVTNDT EVAENNYEAL KDFFRLFPEY KDNKLFLTGE S YAGIYIPT ...String: DRHHHHHHGS APDQDEIDAL PGLAKQPSFR QYSGYLRASD SKHFHYWFVE SQNDPKNSPV VLWLNGGPGC SSLDGLLTEH GPFLIQPDG VTLEYNPYAW NLIANVLYIE SPAGVGFSYS DDKMYVTNDT EVAENNYEAL KDFFRLFPEY KDNKLFLTGE S YAGIYIPT LAVLVMQDPS MNLQGLAVGN GLASYEQNDN SLVYFAYYHG LLGNRLWTSL QTHCCAQNKC NFYDNKDPEC VN NLLEVSR IVGKSGLNIY NLYAPCAGGV PGRHRYEDTL VVQDFGNIFT RLPLKRRFPE ALMRSGDKVR LDPPCTNTTA PSN YLNNPY VRKALHIPES LPRWDMCNFL VNLQYRRLYQ SMNSQYLKLL SSQKYQILLY NGDVDMACNF MGDEWFVDSL NQKM EVQRR PWLVDYGESG EQVAGFVKEC SHITFLTIKG AGHMVPTDKP RAAFTMFSRF LNKEPY UniProtKB: Lysosomal protective protein |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 30 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 4.5 / Details: 50 mM sodium acetate pH 4.5, 50 mM NaCl |
| Grid | Model: Homemade / Material: COPPER/RHODIUM / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | two datasets: untilted and 40 degrees tilted |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 315 / Average electron dose: 41.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 75000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 116 | ||||||
| Output model |  PDB-7kdv: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)