[English] 日本語
 Yorodumi
Yorodumi- EMDB-22797: Nucleosome isolated from interphase chromosome formed in Xenopus ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22797 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nucleosome isolated from interphase chromosome formed in Xenopus egg extract (mono fraction) | |||||||||
 Map data Map data | Fliped and rescaled map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationstructural constituent of chromatin / heterochromatin formation / nucleosome / nucleosome assembly / protein heterodimerization activity / DNA binding / nucleoplasm / nucleus Similarity search - Function | |||||||||
| Biological species | ||||||||||
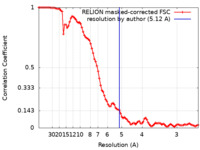
| Method | single particle reconstruction / cryo EM / Resolution: 5.12 Å | |||||||||
 Authors Authors | Arimura Y / Funabiki H | |||||||||
| Funding support |  United States, United States,  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: Structural features of nucleosomes in interphase and metaphase chromosomes. Authors: Yasuhiro Arimura / Rochelle M Shih / Ruby Froom / Hironori Funabiki /  Abstract: Structural heterogeneity of nucleosomes in functional chromosomes is unknown. Here, we devise the template-, reference- and selection-free (TRSF) cryo-EM pipeline to simultaneously reconstruct cryo- ...Structural heterogeneity of nucleosomes in functional chromosomes is unknown. Here, we devise the template-, reference- and selection-free (TRSF) cryo-EM pipeline to simultaneously reconstruct cryo-EM structures of protein complexes from interphase or metaphase chromosomes. The reconstructed interphase and metaphase nucleosome structures are on average indistinguishable from canonical nucleosome structures, despite DNA sequence heterogeneity, cell-cycle-specific posttranslational modifications, and interacting proteins. Nucleosome structures determined by a decoy-classifying method and structure variability analyses reveal the nucleosome structural variations in linker DNA, histone tails, and nucleosome core particle configurations, suggesting that the opening of linker DNA, which is correlated with H2A C-terminal tail positioning, is suppressed in chromosomes. High-resolution (3.4-3.5 Å) nucleosome structures indicate DNA-sequence-independent stabilization of superhelical locations ±0-1 and ±3.5-4.5. The linker histone H1.8 preferentially binds to metaphase chromatin, from which chromatosome cryo-EM structures with H1.8 at the on-dyad position are reconstituted. This study presents the structural characteristics of nucleosomes in chromosomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22797.map.gz emd_22797.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22797-v30.xml emd-22797-v30.xml emd-22797.xml emd-22797.xml | 31.7 KB 31.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22797_fsc.xml emd_22797_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_22797.png emd_22797.png | 29.3 KB | ||
| Others |  emd_22797_additional_1.map.gz emd_22797_additional_1.map.gz emd_22797_additional_2.map.gz emd_22797_additional_2.map.gz emd_22797_additional_3.map.gz emd_22797_additional_3.map.gz emd_22797_additional_4.map.gz emd_22797_additional_4.map.gz emd_22797_additional_5.map.gz emd_22797_additional_5.map.gz emd_22797_additional_6.map.gz emd_22797_additional_6.map.gz emd_22797_additional_7.map.gz emd_22797_additional_7.map.gz emd_22797_half_map_1.map.gz emd_22797_half_map_1.map.gz emd_22797_half_map_2.map.gz emd_22797_half_map_2.map.gz | 4 MB 4 MB 4 MB 4 MB 4 MB 4 MB 59.8 MB 49.8 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22797 http://ftp.pdbj.org/pub/emdb/structures/EMD-22797 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22797 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22797 | HTTPS FTP |
-Related structure data
| Related structure data |  7kbdC  7kbeC  7kbfC  22799  22802 C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10742 (Title: Mono nucleosome fraction from interphase chromosome in Xenopus egg extract lot1 EMPIAR-10742 (Title: Mono nucleosome fraction from interphase chromosome in Xenopus egg extract lot1Data size: 749.0 Data #1: mono nucleosome fraction from interphase chromosome in Xenopus egg extract lot1 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22797.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22797.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Fliped and rescaled map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: 3D classification output map for structural variation analysis
| File | emd_22797_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D classification output map for structural variation analysis | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3D classification output map for structural variation analysis
| File | emd_22797_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D classification output map for structural variation analysis | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3D classification output map for structural variation analysis
| File | emd_22797_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D classification output map for structural variation analysis | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3D classification output map for structural variation analysis
| File | emd_22797_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D classification output map for structural variation analysis | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3D classification output map for structural variation analysis
| File | emd_22797_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D classification output map for structural variation analysis | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3D classification output map for structural variation analysis
| File | emd_22797_additional_6.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D classification output map for structural variation analysis | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: The map before flipping
| File | emd_22797_additional_7.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The map before flipping | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_22797_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_22797_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nucleosome in interphase chromosome formed in Xenopus egg extract...
| Entire | Name: Nucleosome in interphase chromosome formed in Xenopus egg extract (oligo fraction) |
|---|---|
| Components |
|
-Supramolecule #1: Nucleosome in interphase chromosome formed in Xenopus egg extract...
| Supramolecule | Name: Nucleosome in interphase chromosome formed in Xenopus egg extract (oligo fraction) type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 45.02 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)