[English] 日本語
 Yorodumi
Yorodumi- EMDB-20761: Katanin hexamer in the spiral conformation in complex with substrate -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20761 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Katanin hexamer in the spiral conformation in complex with substrate | |||||||||
 Map data Map data | Katanin hexamer in the spiral conformation in complex with substrate | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | katanin / microtubule-severing / MEI-1 / microtubule cytoskeleton / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of meiotic spindle elongation / MATH domain binding / striated muscle myosin thick filament assembly / meiotic spindle disassembly / katanin complex / meiotic spindle pole / microtubule-severing ATPase / microtubule severing ATPase activity / microtubule severing / female meiotic nuclear division ...negative regulation of meiotic spindle elongation / MATH domain binding / striated muscle myosin thick filament assembly / meiotic spindle disassembly / katanin complex / meiotic spindle pole / microtubule-severing ATPase / microtubule severing ATPase activity / microtubule severing / female meiotic nuclear division / meiotic spindle organization / embryo development ending in birth or egg hatching / meiotic spindle / microtubule depolymerization / spindle / spindle pole / protein phosphatase binding / microtubule binding / microtubule / molecular adaptor activity / cell division / centrosome / chromatin / ATP hydrolysis activity / ATP binding / identical protein binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Zehr EA / Roll-Mecak A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Dev Cell / Year: 2020 Journal: Dev Cell / Year: 2020Title: Katanin Grips the β-Tubulin Tail through an Electropositive Double Spiral to Sever Microtubules. Authors: Elena A Zehr / Agnieszka Szyk / Ewa Szczesna / Antonina Roll-Mecak /  Abstract: The AAA ATPase katanin severs microtubules. It is critical in cell division, centriole biogenesis, and neuronal morphogenesis. Its mutation causes microcephaly. The microtubule templates katanin ...The AAA ATPase katanin severs microtubules. It is critical in cell division, centriole biogenesis, and neuronal morphogenesis. Its mutation causes microcephaly. The microtubule templates katanin hexamerization and activates its ATPase. The structural basis for these activities and how they lead to severing is unknown. Here, we show that β-tubulin tails are necessary and sufficient for severing. Cryoelectron microscopy (cryo-EM) structures reveal the essential tubulin tail glutamates gripped by a double spiral of electropositive loops lining the katanin central pore. Each spiral couples allosterically to the ATPase and binds alternating, successive substrate residues, with consecutive residues coordinated by adjacent protomers. This tightly couples tail binding, hexamerization, and ATPase activation. Hexamer structures in different states suggest an ATPase-driven, ratchet-like translocation of the tubulin tail through the pore. A disordered region outside the AAA core anchors katanin to the microtubule while the AAA motor exerts the forces that extract tubulin dimers and sever the microtubule. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20761.map.gz emd_20761.map.gz | 39.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20761-v30.xml emd-20761-v30.xml emd-20761.xml emd-20761.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20761_fsc.xml emd_20761_fsc.xml | 7.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_20761.png emd_20761.png | 197 KB | ||
| Filedesc metadata |  emd-20761.cif.gz emd-20761.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20761 http://ftp.pdbj.org/pub/emdb/structures/EMD-20761 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20761 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20761 | HTTPS FTP |
-Related structure data
| Related structure data |  6ugdMC  6ugeC  6ugfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20761.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20761.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Katanin hexamer in the spiral conformation in complex with substrate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
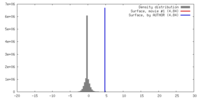
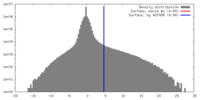
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Hexameric complex of C.elegans katanin bound to polyglutamate peptide
| Entire | Name: Hexameric complex of C.elegans katanin bound to polyglutamate peptide |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric complex of C.elegans katanin bound to polyglutamate peptide
| Supramolecule | Name: Hexameric complex of C.elegans katanin bound to polyglutamate peptide type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 312 KDa |
-Macromolecule #1: Meiotic spindle formation protein mei-1
| Macromolecule | Name: Meiotic spindle formation protein mei-1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: microtubule-severing ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.576336 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSFTMASMTG GQQMGRGSMN GDVQSVIRGY LERAQVAKTM SDAGRWNEAG DLLRQLMTDV KSCKISASNR DEHDARNTFL RALEANLKL VQQNVRDEDD LHEAMTRQSG SPEPPADPDV WSKPSPPLPS SSKFGATKKG VGAAGPRPRE ISKSTSSMST N PADVKPAN ...String: GSFTMASMTG GQQMGRGSMN GDVQSVIRGY LERAQVAKTM SDAGRWNEAG DLLRQLMTDV KSCKISASNR DEHDARNTFL RALEANLKL VQQNVRDEDD LHEAMTRQSG SPEPPADPDV WSKPSPPLPS SSKFGATKKG VGAAGPRPRE ISKSTSSMST N PADVKPAN PTQGILPQNS AGDSFDASAY DAYIVQAVRG TMATNTENTM SLDDIIGMHD VKQVLHEAVT LPLLVPEFFQ GL RSPWKAM VLAGPPGTGK TLIARAIASE SSSTFFTVSS TDLSSKWRGD SEKIVRLLFE LARFYAPSII FIDQIDTLGG QRG NSGEHE ASRRVKSEFL VQMDGSQNKF DSRRVFVLAA TNIPWELDEA LRRRFEKRIF IPLPDIDARK KLIEKSMEGT PKSD EINYD DLAARTEGFS GADVVSLCRT AAINVLRRYD TKSLRGGELT AAMESLKAEL VRNIDFEAAL QAVSPSAGPD TMLKC KEWC DSFGAM UniProtKB: Meiotic spindle formation protein mei-1 |
-Macromolecule #2: Polyglutamate peptide
| Macromolecule | Name: Polyglutamate peptide / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 1.825609 KDa |
| Sequence | String: EEEEEEEEEE EEEE |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 6 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: C-flat-2/1 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 279.15 K / Instrument: LEICA EM GP / Details: Load 5 ul sample, wait 10 sec, blot 4.5 sec. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 93.0 K / Max: 103.0 K |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7420 pixel / Digitization - Dimensions - Height: 7676 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 4 / Number real images: 5911 / Average exposure time: 10.0 sec. / Average electron dose: 73.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.8000000000000003 µm / Calibrated defocus min: 1.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)