[English] 日本語
 Yorodumi
Yorodumi- EMDB-22739: CryoEM reconstruction of SARS-CoV-2 Spike in complex with the Fab... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22739 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM reconstruction of SARS-CoV-2 Spike in complex with the Fab fragment of neutralizing antibody 80. | ||||||||||||||||||
 Map data Map data | CryoEM reconstruction of SARS-CoV-2 Spike in complex with the Fab fragment of neutralizing antibody 80 | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.2 Å | ||||||||||||||||||
 Authors Authors | Kucharska I / Tan YZ / Benlekbir S / Rubinstein JL / Julien JP | ||||||||||||||||||
| Funding support |  Canada, 5 items Canada, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers. Authors: Edurne Rujas / Iga Kucharska / Yong Zi Tan / Samir Benlekbir / Hong Cui / Tiantian Zhao / Gregory A Wasney / Patrick Budylowski / Furkan Guvenc / Jocelyn C Newton / Taylor Sicard / Anthony ...Authors: Edurne Rujas / Iga Kucharska / Yong Zi Tan / Samir Benlekbir / Hong Cui / Tiantian Zhao / Gregory A Wasney / Patrick Budylowski / Furkan Guvenc / Jocelyn C Newton / Taylor Sicard / Anthony Semesi / Krithika Muthuraman / Amy Nouanesengsy / Clare Burn Aschner / Katherine Prieto / Stephanie A Bueler / Sawsan Youssef / Sindy Liao-Chan / Jacob Glanville / Natasha Christie-Holmes / Samira Mubareka / Scott D Gray-Owen / John L Rubinstein / Bebhinn Treanor / Jean-Philippe Julien /    Abstract: SARS-CoV-2, the virus responsible for COVID-19, has caused a global pandemic. Antibodies can be powerful biotherapeutics to fight viral infections. Here, we use the human apoferritin protomer as a ...SARS-CoV-2, the virus responsible for COVID-19, has caused a global pandemic. Antibodies can be powerful biotherapeutics to fight viral infections. Here, we use the human apoferritin protomer as a modular subunit to drive oligomerization of antibody fragments and transform antibodies targeting SARS-CoV-2 into exceptionally potent neutralizers. Using this platform, half-maximal inhibitory concentration (IC) values as low as 9 × 10 M are achieved as a result of up to 10,000-fold potency enhancements compared to corresponding IgGs. Combination of three different antibody specificities and the fragment crystallizable (Fc) domain on a single multivalent molecule conferred the ability to overcome viral sequence variability together with outstanding potency and IgG-like bioavailability. The MULTi-specific, multi-Affinity antiBODY (Multabody or MB) platform thus uniquely leverages binding avidity together with multi-specificity to deliver ultrapotent and broad neutralizers against SARS-CoV-2. The modularity of the platform also makes it relevant for rapid evaluation against other infectious diseases of global health importance. Neutralizing antibodies are a promising therapeutic for SARS-CoV-2. #1:  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: Multivalency transforms SARS-CoV-2 antibodies into broad and ultrapotent neutralizers Authors: Rujas E / Kucharska I / Tan YZ / Benlekbir S / Cui H / Zhao T / Wasney G / Budylowski P / Guvenc F / Newton JC / Sicard T / Semesi A / Muthuraman K / Nouanesengsy A / Prieto K / Bueler SA / ...Authors: Rujas E / Kucharska I / Tan YZ / Benlekbir S / Cui H / Zhao T / Wasney G / Budylowski P / Guvenc F / Newton JC / Sicard T / Semesi A / Muthuraman K / Nouanesengsy A / Prieto K / Bueler SA / Youssef S / Liao-Chan S / Glanville J / Christie-Holmes N / Mubareka S / Gray-Owen SD / Rubinstein JL / Treanor B / Julien JP | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22739.map.gz emd_22739.map.gz | 49.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22739-v30.xml emd-22739-v30.xml emd-22739.xml emd-22739.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22739.png emd_22739.png | 98 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22739 http://ftp.pdbj.org/pub/emdb/structures/EMD-22739 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22739 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22739 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22739.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22739.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM reconstruction of SARS-CoV-2 Spike in complex with the Fab fragment of neutralizing antibody 80 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.50208 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
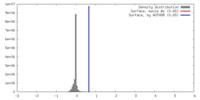
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SARS-CoV-2 Spike in complex with the Fab fragment of neutralizing...
| Entire | Name: SARS-CoV-2 Spike in complex with the Fab fragment of neutralizing antibody 80 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Spike in complex with the Fab fragment of neutralizing...
| Supramolecule | Name: SARS-CoV-2 Spike in complex with the Fab fragment of neutralizing antibody 80 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Fab fragment of neutralizing antibody 80
| Supramolecule | Name: Fab fragment of neutralizing antibody 80 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: SARS-CoV-2 Spike
| Supramolecule | Name: SARS-CoV-2 Spike / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Fab 80 heavy chain
| Macromolecule | Name: Fab 80 heavy chain / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGSSVKV SCKASGGTFN RYAFSWVRQA PGQGLEWMGG IIPIFGTANY AQKFQGRVTI TADESTSTAY MELSSLRSED TAVYYCARST RELPEVVDWY FDLWGRGTLV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP VTVSWNSGA ...String: QVQLVQSGAE VKKPGSSVKV SCKASGGTFN RYAFSWVRQA PGQGLEWMGG IIPIFGTANY AQKFQGRVTI TADESTSTAY MELSSLRSED TAVYYCARST RELPEVVDWY FDLWGRGTLV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP VTVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV N HKPSNTKV DKKVEPKSC |
-Macromolecule #2: Fab 80 light chain
| Macromolecule | Name: Fab 80 light chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQSPDS LAVSLGERAT INCKSSQSVL YSSNNKNYLA WYQQKPGQPP KLLIYWASTR ESGVPDRFSG SGSGTDFTLT ISSLQAEDVA VYYCQQYYSA PLTFGGGTKV EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPRE AKVQWKVDNA LQSGNSQESV ...String: DIVMTQSPDS LAVSLGERAT INCKSSQSVL YSSNNKNYLA WYQQKPGQPP KLLIYWASTR ESGVPDRFSG SGSGTDFTLT ISSLQAEDVA VYYCQQYYSA PLTFGGGTKV EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPRE AKVQWKVDNA LQSGNSQESV TEQDSKDSTY SLSSTLTLSK ADYEKHKVYA CEVTHQGLS SPVTKSFNRG EC |
-Macromolecule #3: SARS-CoV-2 Spike
| Macromolecule | Name: SARS-CoV-2 Spike / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQL PPAYT NSFTRGVYYP DKVFRSSVLH STQDLFLPFF SNVTWFHAIH VSGTN GTKR FDNPVLPFND GVYFASTEKS NIIRGWIFGT TLDSKTQSLL IVNNAT NVV IKVCEFQFCN DPFLGVYYHK NNKSWMESEF RVYSSANNCT ...String: MFVFLVLLPL VSSQCVNLTT RTQL PPAYT NSFTRGVYYP DKVFRSSVLH STQDLFLPFF SNVTWFHAIH VSGTN GTKR FDNPVLPFND GVYFASTEKS NIIRGWIFGT TLDSKTQSLL IVNNAT NVV IKVCEFQFCN DPFLGVYYHK NNKSWMESEF RVYSSANNCT FEYVSQP FL MDLEGKQGNF KNLREFVFKN IDGYFKIYSK HTPINLVRDL PQGFSALE P LVDLPIGINI TRFQTLLALH RSYLTPGDSS SGWTAGAAAY YVGYLQPRT FLLKYNENGT ITDAVDCALD PLSETKCTLK SFTVEKGIYQ TSNFRVQPTE SIVRFPNIT NLCPFGEVFN ATRFASVYAW NRKRISNCVA DYSVLYNSAS F STFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTGKIADYN YK LPDDFTG CVIAWNSNNL DSKVGGNYNY LYRLFRKSNL KPFERDISTE IYQ AGSTPC NGVEGFNCYF PLQSYGFQPT NGVGYQPYRV VVLSFELLHA PATV CGPKK STNLVKNKCV NFNFNGLTGT GVLTESNKKF LPFQQFGRDI ADTTD AVRD PQTLEILDIT PCSFGGVSVI TPGTNTSNQV AVLYQDVNCT EVPVAI HAD QLTPTWRVYS TGSNVFQTRA GCLIGAEHVN NSYECDIPIG AGICASY QT QTNSPASVAS QSIIAYTMSL GAENSVAYSN NSIAIPTNFT ISVTTEIL P VSMTKTSVDC TMYICGDSTE CSNLLLQYGS FCTQLNRALT GIAVEQDKN TQEVFAQVKQ IYKTPPIKDF GGFNFSQILP DPSKPSKRSF IEDLLFNKVT LADAGFIKQ YGDCLGDIAA RDLICAQKFN GLTVLPPLLT DEMIAQYTSA L LAGTITSG WTFGAGAALQ IPFAMQMAYR FNGIGVTQNV LYENQKLIAN QF NSAIGKI QDSLSSTASA LGKLQDVVNQ NAQALNTLVK QLSSNFGAIS SVL NDILSR LDPPEAEVQI DRLITGRLQS LQTYVTQQLI RAAEIRASAN LAAT KMSEC VLGQSKRVDF CGKGYHLMSF PQSAPHGVVF LHVTYVPAQE KNFTT APAI CHDGKAHFPR EGVFVSNGTH WFVTQRNFYE PQIITTDNTF VSGNCD VVI GIVNNTVYDP LQPELDSFKE ELDKYFKNHT SPDVDLGDIS GINASVV NI QKEIDRLNEV AKNLNESLID LQELGKYEQY IKWPSGRLVP RGSPGSGY I PEAPRDGQAY VRKDGEWVLL STFLGHHHHH H |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Homemade / Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: glow discharged with PELCO easiGlow (Ted Pella) | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 3610 / Average exposure time: 9.6 sec. / Average electron dose: 45.0 e/Å2 Details: 820 images under 0 degrees tilt, 2790 images under 40 degrees tilt |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)