+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22700 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Density of TyTx4 Fab in Complex with Typhoid Toxin | ||||||||||||
 Map data Map data | Full-map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Typhoid Toxin / A2B5 / Antibody / Fab / TOXIN | ||||||||||||
| Function / homology | Bordetella pertussis toxin B, subunit 2/3, C-terminal / Pertussis toxin, subunit 2 and 3, C-terminal domain / Enterotoxin / extracellular region / Pertussis-like toxin subunit Function and homology information Function and homology information | ||||||||||||
| Biological species |  Salmonella enterica subsp. enterica serovar Typhi (bacteria) / Salmonella enterica subsp. enterica serovar Typhi (bacteria) /   Salmonella typhi (bacteria) Salmonella typhi (bacteria) | ||||||||||||
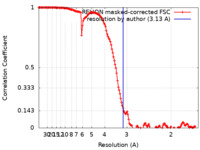
| Method | single particle reconstruction / cryo EM / Resolution: 3.13 Å | ||||||||||||
 Authors Authors | Nguyen T / Feathers JR | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: The structural basis of Salmonella AB toxin neutralization by antibodies targeting the glycan-receptor binding subunits. Authors: Tri Nguyen / Angelene F Richards / Durga P Neupane / J Ryan Feathers / Yi-An Yang / Ji Hyun Sim / Haewon Byun / Sohyoung Lee / Changhwan Ahn / Greta Van Slyke / J Christopher Fromme / ...Authors: Tri Nguyen / Angelene F Richards / Durga P Neupane / J Ryan Feathers / Yi-An Yang / Ji Hyun Sim / Haewon Byun / Sohyoung Lee / Changhwan Ahn / Greta Van Slyke / J Christopher Fromme / Nicholas J Mantis / Jeongmin Song /  Abstract: Many bacterial pathogens secrete AB toxins comprising two functionally distinct yet complementary "A" and "B" subunits to benefit the pathogens during infection. The lectin-like pentameric B subunits ...Many bacterial pathogens secrete AB toxins comprising two functionally distinct yet complementary "A" and "B" subunits to benefit the pathogens during infection. The lectin-like pentameric B subunits recognize specific sets of host glycans to deliver the toxin into target host cells. Here, we offer the molecular mechanism by which neutralizing antibodies, which have the potential to bind to all glycan-receptor binding sites and thus completely inhibit toxin binding to host cells, are inhibited from exerting this action. Cryogenic electron microscopy (cryo-EM)-based analyses indicate that the skewed positioning of the toxin A subunit(s) toward one side of the toxin B pentamer inhibited neutralizing antibody binding to the laterally located epitopes, rendering some glycan-receptor binding sites that remained available for the toxin binding and endocytosis process, which is strikingly different from the counterpart antibodies recognizing the far side-located epitopes. These results highlight additional features of the toxin-antibody interactions and offer important insights into anti-toxin strategies. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22700.map.gz emd_22700.map.gz | 191.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22700-v30.xml emd-22700-v30.xml emd-22700.xml emd-22700.xml | 25.6 KB 25.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22700_fsc.xml emd_22700_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_22700.png emd_22700.png | 51.2 KB | ||
| Filedesc metadata |  emd-22700.cif.gz emd-22700.cif.gz | 6.7 KB | ||
| Others |  emd_22700_additional_1.map.gz emd_22700_additional_1.map.gz emd_22700_additional_2.map.gz emd_22700_additional_2.map.gz emd_22700_half_map_1.map.gz emd_22700_half_map_1.map.gz emd_22700_half_map_2.map.gz emd_22700_half_map_2.map.gz | 227 MB 13.2 MB 193.9 MB 193.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22700 http://ftp.pdbj.org/pub/emdb/structures/EMD-22700 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22700 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22700 | HTTPS FTP |
-Related structure data
| Related structure data |  7k7iMC  7k7hC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22700.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22700.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full-map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Post-processed Map
| File | emd_22700_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
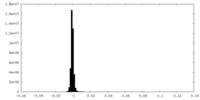
| Density Histograms |
-Additional map: Post-processed Masked Map
| File | emd_22700_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed Masked Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
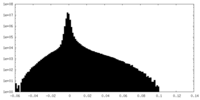
| Density Histograms |
-Half map: Half-map 1
| File | emd_22700_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
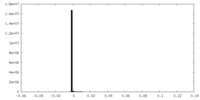
| Density Histograms |
-Half map: Half-map 2
| File | emd_22700_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
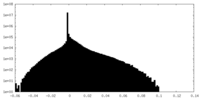
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of TyTx4 Fab with Typhoid Toxin Catalytic Inactivated Mutant
| Entire | Name: Complex of TyTx4 Fab with Typhoid Toxin Catalytic Inactivated Mutant |
|---|---|
| Components |
|
-Supramolecule #1: Complex of TyTx4 Fab with Typhoid Toxin Catalytic Inactivated Mutant
| Supramolecule | Name: Complex of TyTx4 Fab with Typhoid Toxin Catalytic Inactivated Mutant type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Map of Fab fragment of proteolytically cleaved IgG antibody TyTx4 in complex with purified Typhoid toxin PltA E133A and CdtB H160Q double mutant |
|---|
-Supramolecule #2: Typhoid Toxin Catalytically Inactivated Mutant
| Supramolecule | Name: Typhoid Toxin Catalytically Inactivated Mutant / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: S. Typhi A2B5 toxin PltA E133A & CdtB H160Q double mutant |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhi (bacteria) Salmonella enterica subsp. enterica serovar Typhi (bacteria) |
-Supramolecule #3: TyTx4 Fab
| Supramolecule | Name: TyTx4 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 Details: Fab fragment generated by proteolytic cleavage of IgG antibody TyTx4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Putative pertussis-like toxin subunit
| Macromolecule | Name: Putative pertussis-like toxin subunit / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhi (bacteria) Salmonella typhi (bacteria) |
| Molecular weight | Theoretical: 12.563042 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EWTGDNTNAY YSDEVISELH VGQIDTSPYF CIKTVKANGS GTPVVACAVS KQSIWAPSFK ELLDQARYFY STGQSVRIHV QKNIWTYPL FVNTFSANAL VGLSSCSATQ CFGPK UniProtKB: Pertussis-like toxin subunit |
-Macromolecule #2: Fab Light Chain Variable Domain
| Macromolecule | Name: Fab Light Chain Variable Domain / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.753811 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DILLTQSPSI LSVSPGERVS FSCRASQSIG TSIHWYQQKP NGSPRLLIQY ASQSISGIPS RFSGSGSGTD FTLTINSVES EDIADYYCQ HTNGWPYTFG WGDHAGNKP |
-Macromolecule #3: Fab Heavy Chain Variable Domain
| Macromolecule | Name: Fab Heavy Chain Variable Domain / type: protein_or_peptide / ID: 3 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.15971 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VKLVESGGGL VKPGGSLKLS CAASGFAFST YDMSWVRQTP EKRLEWVATI SGGGSYTYYP DIVKGRFTIS RDNARNTLYL QMSSLRSED TALYFCVRQY YGSSNYGMDY WGQGTSVTVS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK I / Details: blot for 2.0 second before plunging. | |||||||||
| Details | Freshly prepared size-exclusion-chromatography purified complex between Fab fragment of TyTx4 and Typhoid toxin wild-type |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 51.5 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 49000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Details | Initial local fitting was done using Chimera and then Coot was used for rebuilding Fab variable domains into correct sequences. Refinement was performed using Real Space Refine in PHENIX and was iterated with manual building in Coot. | ||||||||
| Output model |  PDB-7k7i: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)