[English] 日本語
 Yorodumi
Yorodumi- EMDB-22635: Structure of the Bacterial Ribosome at 2 Angstrom Resolution (30S... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22635 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Bacterial Ribosome at 2 Angstrom Resolution (30S head domain focused refinement) | |||||||||
 Map data Map data | 30S head domain focused refinement | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
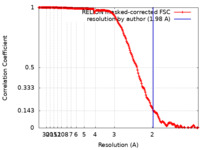
| Method | single particle reconstruction / cryo EM / Resolution: 1.98 Å | |||||||||
 Authors Authors | Watson ZL / Ward FR / Meheust R / Ad O / Schepartz A / Banfield JF / Cate JHD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Structure of the bacterial ribosome at 2 Å resolution. Authors: Zoe L Watson / Fred R Ward / Raphaël Méheust / Omer Ad / Alanna Schepartz / Jillian F Banfield / Jamie Hd Cate /  Abstract: Using cryo-electron microscopy (cryo-EM), we determined the structure of the 70S ribosome with a global resolution of 2.0 Å. The maps reveal unambiguous positioning of protein and RNA residues, ...Using cryo-electron microscopy (cryo-EM), we determined the structure of the 70S ribosome with a global resolution of 2.0 Å. The maps reveal unambiguous positioning of protein and RNA residues, their detailed chemical interactions, and chemical modifications. Notable features include the first examples of isopeptide and thioamide backbone substitutions in ribosomal proteins, the former likely conserved in all domains of life. The maps also reveal extensive solvation of the small (30S) ribosomal subunit, and interactions with A-site and P-site tRNAs, mRNA, and the antibiotic paromomycin. The maps and models of the bacterial ribosome presented here now allow a deeper phylogenetic analysis of ribosomal components including structural conservation to the level of solvation. The high quality of the maps should enable future structural analyses of the chemical basis for translation and aid the development of robust tools for cryo-EM structure modeling and refinement. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22635.map.gz emd_22635.map.gz | 436.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22635-v30.xml emd-22635-v30.xml emd-22635.xml emd-22635.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22635_fsc.xml emd_22635_fsc.xml | 17.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_22635.png emd_22635.png | 82.8 KB | ||
| Others |  emd_22635_half_map_1.map.gz emd_22635_half_map_1.map.gz emd_22635_half_map_2.map.gz emd_22635_half_map_2.map.gz | 373.6 MB 373.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22635 http://ftp.pdbj.org/pub/emdb/structures/EMD-22635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22635 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22635.map.gz / Format: CCP4 / Size: 465.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22635.map.gz / Format: CCP4 / Size: 465.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 30S head domain focused refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.7118 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: 30S head domain focused refined half map 1
| File | emd_22635_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 30S head domain focused refined half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
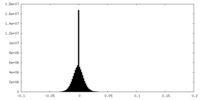
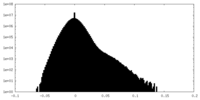
| Density Histograms |
-Half map: 30S head domain focused refined half map 2
| File | emd_22635_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 30S head domain focused refined half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
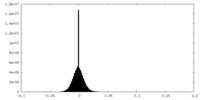
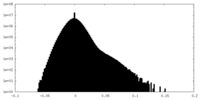
| Density Histograms |
- Sample components
Sample components
-Entire : E. coli 70S ribosome
| Entire | Name: E. coli 70S ribosome |
|---|---|
| Components |
|
-Supramolecule #1: E. coli 70S ribosome
| Supramolecule | Name: E. coli 70S ribosome / type: complex / ID: 1 / Parent: 0 Details: E. coli 70S ribosome bound to mRNA, tRNAs, and paromomycin |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.7 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.27 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)