+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22528 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Succinate: quinone oxidoreductase SQR from E.coli K12 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Succinate: quinone oxidoreductase / SdhA / sdhB / SdhC / SdhD / ELECTRON TRANSPORT / ELECTRON TRANSPORT-OXIDOREDUCTASE complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsuccinate dehydrogenase activity / respiratory chain complex II (succinate dehydrogenase) / succinate dehydrogenase (quinone) activity / succinate dehydrogenase / cytochrome complex assembly / aerobic electron transport chain / anaerobic respiration / 3 iron, 4 sulfur cluster binding / ubiquinone binding / iron-sulfur cluster binding ...succinate dehydrogenase activity / respiratory chain complex II (succinate dehydrogenase) / succinate dehydrogenase (quinone) activity / succinate dehydrogenase / cytochrome complex assembly / aerobic electron transport chain / anaerobic respiration / 3 iron, 4 sulfur cluster binding / ubiquinone binding / iron-sulfur cluster binding / membrane => GO:0016020 / tricarboxylic acid cycle / aerobic respiration / respiratory electron transport chain / 2 iron, 2 sulfur cluster binding / flavin adenine dinucleotide binding / 4 iron, 4 sulfur cluster binding / electron transfer activity / heme binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Lyu M / Su C-C | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2021 Journal: Nat Methods / Year: 2021Title: A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins. Authors: Chih-Chia Su / Meinan Lyu / Christopher E Morgan / Jani Reddy Bolla / Carol V Robinson / Edward W Yu /   Abstract: Single-particle cryo-electron microscopy (cryo-EM) has become a powerful technique in the field of structural biology. However, the inability to reliably produce pure, homogeneous membrane protein ...Single-particle cryo-electron microscopy (cryo-EM) has become a powerful technique in the field of structural biology. However, the inability to reliably produce pure, homogeneous membrane protein samples hampers the progress of their structural determination. Here, we develop a bottom-up iterative method, Build and Retrieve (BaR), that enables the identification and determination of cryo-EM structures of a variety of inner and outer membrane proteins, including membrane protein complexes of different sizes and dimensions, from a heterogeneous, impure protein sample. We also use the BaR methodology to elucidate structural information from Escherichia coli K12 crude membrane and raw lysate. The findings demonstrate that it is possible to solve high-resolution structures of a number of relatively small (<100 kDa) and less abundant (<10%) unidentified membrane proteins within a single, heterogeneous sample. Importantly, these results highlight the potential of cryo-EM for systems structural proteomics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22528.map.gz emd_22528.map.gz | 9.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22528-v30.xml emd-22528-v30.xml emd-22528.xml emd-22528.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22528.png emd_22528.png | 279 KB | ||
| Filedesc metadata |  emd-22528.cif.gz emd-22528.cif.gz | 6.1 KB | ||
| Others |  emd_22528_additional_1.map.gz emd_22528_additional_1.map.gz | 154.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22528 http://ftp.pdbj.org/pub/emdb/structures/EMD-22528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22528 | HTTPS FTP |
-Related structure data
| Related structure data |  7jz2MC  6wtiC  6wtzC  6wu0C  6wu6C  7jz3C  7jz6C  7jzhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22528.map.gz / Format: CCP4 / Size: 10.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22528.map.gz / Format: CCP4 / Size: 10.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
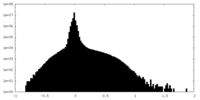
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_22528_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
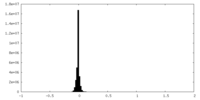
| Density Histograms |
- Sample components
Sample components
+Entire : Succinate: quinone oxidoreductase SQR from E.coli K12
+Supramolecule #1: Succinate: quinone oxidoreductase SQR from E.coli K12
+Macromolecule #1: Succinate dehydrogenase flavoprotein subunit
+Macromolecule #2: Succinate dehydrogenase iron-sulfur subunit
+Macromolecule #3: Succinate dehydrogenase cytochrome b556 subunit
+Macromolecule #4: Succinate dehydrogenase hydrophobic membrane anchor subunit
+Macromolecule #5: FLAVIN-ADENINE DINUCLEOTIDE
+Macromolecule #6: SODIUM ION
+Macromolecule #7: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #8: IRON/SULFUR CLUSTER
+Macromolecule #9: FE3-S4 CLUSTER
+Macromolecule #10: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #11: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #12: UBIQUINONE-2
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 38471 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller





























 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)