[English] 日本語
 Yorodumi
Yorodumi- EMDB-22364: Helical filaments of plant light-dependent protochlorophyllide ox... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22364 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical filaments of plant light-dependent protochlorophyllide oxidoreductase (LPOR) bound to NADPH, Pchlide, and membrane | ||||||||||||
 Map data Map data | RELION sharpened map (B factor -97) with local symmetry and then helical symmetry applied to the entire map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | reductase / light-activated / ligand-protein complex / PHOTOSYNTHESIS | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationprotochlorophyllide reductase / protochlorophyllide reductase activity / response to ethylene / chlorophyll biosynthetic process / chloroplast outer membrane / chloroplast thylakoid / chloroplast envelope / chloroplast thylakoid membrane / photosynthesis / chloroplast ...protochlorophyllide reductase / protochlorophyllide reductase activity / response to ethylene / chlorophyll biosynthetic process / chloroplast outer membrane / chloroplast thylakoid / chloroplast envelope / chloroplast thylakoid membrane / photosynthesis / chloroplast / protein domain specific binding / mRNA binding / cytosol Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
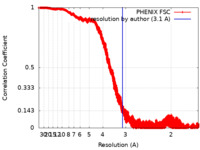
| Method | helical reconstruction / cryo EM / Resolution: 3.1 Å | ||||||||||||
 Authors Authors | Nguyen HC / Gabruk M | ||||||||||||
| Funding support |  Poland, Poland,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2021 Journal: Nat Plants / Year: 2021Title: Photocatalytic LPOR forms helical lattices that shape membranes for chlorophyll synthesis. Authors: Henry C Nguyen / Arthur A Melo / Jerzy Kruk / Adam Frost / Michal Gabruk /   Abstract: Chlorophyll biosynthesis, crucial to life on Earth, is tightly regulated because its precursors are phototoxic. In flowering plants, the enzyme light-dependent protochlorophyllide oxidoreductase ...Chlorophyll biosynthesis, crucial to life on Earth, is tightly regulated because its precursors are phototoxic. In flowering plants, the enzyme light-dependent protochlorophyllide oxidoreductase (LPOR) captures photons to catalyse the penultimate reaction: the reduction of a double bond within protochlorophyllide (Pchlide) to generate chlorophyllide (Chlide). In darkness, LPOR oligomerizes to facilitate photon energy transfer and catalysis. However, the complete three-dimensional structure of LPOR, the higher-order architecture of LPOR oligomers and the implications of these self-assembled states for catalysis, including how LPOR positions Pchlide and the co-factor NADPH, remain unknown. Here, we report the atomic structure of LPOR assemblies by electron cryo-microscopy. LPOR polymerizes with its substrates into helical filaments around constricted lipid bilayer tubes. Portions of LPOR and Pchlide insert into the outer membrane leaflet, targeting the product, Chlide, to the membrane for the final reaction site of chlorophyll biosynthesis. In addition to its crucial photocatalytic role, we show that in darkness LPOR filaments directly shape membranes into high-curvature tubules with the spectral properties of the prolamellar body, whose light-triggered disassembly provides lipids for thylakoid assembly. Moreover, our structure of the catalytic site challenges previously proposed reaction mechanisms. Together, our results reveal a new and unexpected synergy between photosynthetic membrane biogenesis and chlorophyll synthesis in plants, orchestrated by LPOR. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22364.map.gz emd_22364.map.gz | 48.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22364-v30.xml emd-22364-v30.xml emd-22364.xml emd-22364.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22364_fsc.xml emd_22364_fsc.xml | 3.5 MB | Display |  FSC data file FSC data file |
| Images |  emd_22364.png emd_22364.png | 468.7 KB | ||
| Masks |  emd_22364_msk_1.map emd_22364_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22364.cif.gz emd-22364.cif.gz | 6.1 KB | ||
| Others |  emd_22364_additional_1.map.gz emd_22364_additional_1.map.gz emd_22364_additional_2.map.gz emd_22364_additional_2.map.gz emd_22364_half_map_1.map.gz emd_22364_half_map_1.map.gz emd_22364_half_map_2.map.gz emd_22364_half_map_2.map.gz | 200.3 MB 170.9 MB 170.8 MB 170.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22364 http://ftp.pdbj.org/pub/emdb/structures/EMD-22364 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22364 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22364 | HTTPS FTP |
-Related structure data
| Related structure data |  7jk9MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22364.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22364.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION sharpened map (B factor -97) with local symmetry and then helical symmetry applied to the entire map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.11333 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
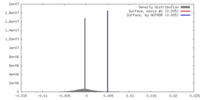
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
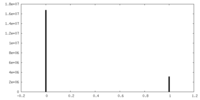
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22364_msk_1.map emd_22364_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
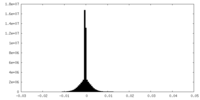
| Density Histograms |
-Additional map: RELION sharpened map (B factor -97) with central Z of 40%
| File | emd_22364_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION sharpened map (B factor -97) with central Z of 40% | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: RELION Refine3D summed and filtered map with central Z of 40%
| File | emd_22364_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION Refine3D summed and filtered map with central Z of 40% | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: RELION Refine3D unfiltered half map with central Z of 40%
| File | emd_22364_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION Refine3D unfiltered half map with central Z of 40% | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: RELION Refine3D unfiltered half map with central Z of 40%
| File | emd_22364_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION Refine3D unfiltered half map with central Z of 40% | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : light-dependent protochlorophyllide oxidoreductase bound to NADPH...
| Entire | Name: light-dependent protochlorophyllide oxidoreductase bound to NADPH, Pchlide, and lipid membrane |
|---|---|
| Components |
|
-Supramolecule #1: light-dependent protochlorophyllide oxidoreductase bound to NADPH...
| Supramolecule | Name: light-dependent protochlorophyllide oxidoreductase bound to NADPH, Pchlide, and lipid membrane type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Protochlorophyllide reductase B, chloroplastic
| Macromolecule | Name: Protochlorophyllide reductase B, chloroplastic / type: protein_or_peptide / ID: 1 / Number of copies: 40 / Enantiomer: LEVO / EC number: protochlorophyllide reductase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.415199 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MALQAASLVS SAFSVRKDAK LNASSSSFKD SSLFGASITD QIKSEHGSSS LRFKREQSLR NLAIRAQTAA TSSPTVTKSV DGKKTLRKG NVVVTGASSG LGLATAKALA ETGKWNVIMA CRDFLKAERA AKSVGMPKDS YTVMHLDLAS LDSVRQFVDN F RRTETPLD ...String: MALQAASLVS SAFSVRKDAK LNASSSSFKD SSLFGASITD QIKSEHGSSS LRFKREQSLR NLAIRAQTAA TSSPTVTKSV DGKKTLRKG NVVVTGASSG LGLATAKALA ETGKWNVIMA CRDFLKAERA AKSVGMPKDS YTVMHLDLAS LDSVRQFVDN F RRTETPLD VLVCNAAVYF PTAKEPTYSA EGFELSVATN HLGHFLLARL LLDDLKKSDY PSKRLIIVGS ITGNTNTLAG NV PPKANLG DLRGLAGGLN GLNSSAMIDG GDFDGAKAYK DSKVCNMLTM QEFHRRFHEE TGVTFASLYP GCIASTGLFR EHI PLFRAL FPPFQKYITK GYVSETESGK RLAQVVSDPS LTKSGVYWSW NNASASFENQ LSEEASDVEK ARKVWEISEK LVGL A UniProtKB: Protochlorophyllide reductase B, chloroplastic |
-Macromolecule #2: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
| Macromolecule | Name: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE type: ligand / ID: 2 / Number of copies: 40 / Formula: NDP |
|---|---|
| Molecular weight | Theoretical: 745.421 Da |
| Chemical component information |  ChemComp-NDP: |
-Macromolecule #3: Protochlorophyllide
| Macromolecule | Name: Protochlorophyllide / type: ligand / ID: 3 / Number of copies: 40 / Formula: PMR |
|---|---|
| Molecular weight | Theoretical: 612.957 Da |
| Chemical component information |  ChemComp-PMR: |
-Macromolecule #4: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
| Macromolecule | Name: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE / type: ligand / ID: 4 / Number of copies: 40 / Formula: LMG |
|---|---|
| Molecular weight | Theoretical: 787.158 Da |
| Chemical component information |  ChemComp-LMG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.1 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 73.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)