+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1760 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Double filaments of C-terminally GFP-fused TubZ | |||||||||
 Map data Map data | Double helical filaments of a C-Terminal fusion of GFPmut2 to Bacillus thuringiensis serovar israelensis (ATCC 35646) TubZ (Q8KNP3_BACTI) Density for the GFP fusion of TubZ to remove positive density on the outer boundary of the negative stain and unconnected to the main helix. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cytoskeleton / DNA segregation / FtsZ / FtsZ-like / pBtoxis / pbt156 / plasmid partitioning / RepX / tubulin / tubulin-like / TubZ | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / negative staining / Resolution: 35.0 Å | |||||||||
 Authors Authors | Aylett CHS / Amos LA / Lowe J | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2010 Journal: Proc Natl Acad Sci U S A / Year: 2010Title: Filament structure of bacterial tubulin homologue TubZ. Authors: Christopher H S Aylett / Qing Wang / Katharine A Michie / Linda A Amos / Jan Löwe /  Abstract: Low copy number plasmids often depend on accurate partitioning systems for their continued survival. Generally, such systems consist of a centromere-like region of DNA, a DNA-binding adaptor, and a ...Low copy number plasmids often depend on accurate partitioning systems for their continued survival. Generally, such systems consist of a centromere-like region of DNA, a DNA-binding adaptor, and a polymerizing cytomotive filament. Together these components drive newly replicated plasmids to opposite ends of the dividing cell. The Bacillus thuringiensis plasmid pBToxis relies on a filament of the tubulin/FtsZ-like protein TubZ for its segregation. By combining crystallography and electron microscopy, we have determined the structure of this filament. We explain how GTP hydrolysis weakens the subunit-subunit contact and also shed light on the partitioning of the plasmid-adaptor complex. The double helical superstructure of TubZ filaments is unusual for tubulin-like proteins. Filaments of ParM, the actin-like partitioning protein, are also double helical. We suggest that convergent evolution shapes these different types of cytomotive filaments toward a general mechanism for plasmid separation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1760.map.gz emd_1760.map.gz | 80.5 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1760-v30.xml emd-1760-v30.xml emd-1760.xml emd-1760.xml | 8.5 KB 8.5 KB | Display Display |  EMDB header EMDB header |
| Images |  1760.png 1760.png | 435.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1760 http://ftp.pdbj.org/pub/emdb/structures/EMD-1760 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1760 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1760 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1760.map.gz / Format: CCP4 / Size: 666 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1760.map.gz / Format: CCP4 / Size: 666 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Double helical filaments of a C-Terminal fusion of GFPmut2 to Bacillus thuringiensis serovar israelensis (ATCC 35646) TubZ (Q8KNP3_BACTI) Density for the GFP fusion of TubZ to remove positive density on the outer boundary of the negative stain and unconnected to the main helix. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X: 4.4 Å / Y: 4.4 Å / Z: 4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Double filaments of TubZ-GFP
| Entire | Name: Double filaments of TubZ-GFP |
|---|---|
| Components |
|
-Supramolecule #1000: Double filaments of TubZ-GFP
| Supramolecule | Name: Double filaments of TubZ-GFP / type: sample / ID: 1000 / Details: Negatively stained / Oligomeric state: Dimer / Number unique components: 1 |
|---|
-Macromolecule #1: Cytomotive filament
| Macromolecule | Name: Cytomotive filament / type: protein_or_peptide / ID: 1 / Name.synonym: Cytomotive filament / Oligomeric state: Double filament / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 50 mM NaHEPES 7.5 150 mM KCl 5 mM MgCl2 1 mM GTPyS |
| Staining | Type: NEGATIVE / Details: 1% Uranyl Acetate |
| Grid | Details: CuRh 300 mesh |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Category: CCD / Film or detector model: KODAK SO-163 FILM / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 67000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Axial symmetry: C2 (2 fold cyclic) Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 35.0 Å / Resolution method: OTHER / Software - Name: MRC Details: Final map was calculated from two averaged filaments |
|---|
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera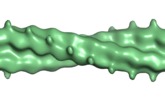








 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)





















