[English] 日本語
 Yorodumi
Yorodumi- EMDB-22341: CryoEM structure of Streptococcus thermophilus SHP pheromone rece... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22341 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Streptococcus thermophilus SHP pheromone receptor Rgg3 in complex with SHP3 | |||||||||
 Map data Map data | Sharpened map generated by cryoSPARC | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA binding transcription factor / quorum sensing / RRNPP / pheromone binding / DNA BINDING PROTEIN | |||||||||
| Function / homology | Transcription activator MutR, C-terminal / HTH-type transcriptional regulator Rgg, C-terminal domain / : / Cro/C1-type HTH domain profile. / Cro/C1-type helix-turn-helix domain / Lambda repressor-like, DNA-binding domain superfamily / Tetratricopeptide-like helical domain superfamily / DNA binding / Positive transcriptional regulator MutR family Function and homology information Function and homology information | |||||||||
| Biological species |  Streptococcus thermophilus (bacteria) / Streptococcus thermophilus (bacteria) /  Streptococcus thermophilus (strain ATCC BAA-250 / LMG 18311) (bacteria) / Streptococcus thermophilus (strain ATCC BAA-250 / LMG 18311) (bacteria) /  Streptococcus thermophilus CNRZ1066 (bacteria) Streptococcus thermophilus CNRZ1066 (bacteria) | |||||||||
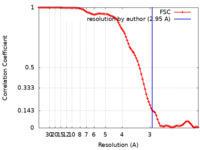
| Method | single particle reconstruction / cryo EM / Resolution: 2.95 Å | |||||||||
 Authors Authors | Petrou VI / Capodagli GC | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Structure-function studies of Rgg binding to pheromones and target promoters reveal a model of transcription factor interplay. Authors: Glenn C Capodagli / Kaitlyn M Tylor / Jason T Kaelber / Vasileios I Petrou / Michael J Federle / Matthew B Neiditch /  Abstract: Regulator gene of glucosyltransferase (Rgg) family proteins, such as Rgg2 and Rgg3, have emerged as primary quorum-sensing regulated transcription factors in species, controlling virulence, ...Regulator gene of glucosyltransferase (Rgg) family proteins, such as Rgg2 and Rgg3, have emerged as primary quorum-sensing regulated transcription factors in species, controlling virulence, antimicrobial resistance, and biofilm formation. Rgg2 and Rgg3 function is regulated by their interaction with oligopeptide quorum-sensing signals called short hydrophobic peptides (SHPs). The molecular basis of Rgg-SHP and Rgg-target DNA promoter specificity was unknown. To close this gap, we determined the cryoelectron microscopy (cryo-EM) structure of Rgg3 bound to its quorum-sensing signal, SHP3, and the X-ray crystal structure of Rgg3 alone. Comparison of these structures with that of an Rgg in complex with cyclosporin A (CsA), an inhibitor of SHP-induced Rgg activity, reveals the molecular basis of CsA function. Furthermore, to determine how Rgg proteins recognize DNA promoters, we determined X-ray crystal structures of both Rgg2 and Rgg3 in complex with their target DNA promoters. The physiological importance of observed Rgg-DNA interactions was dissected using in vivo genetic experiments and in vitro biochemical assays. Based on these structure-function studies, we present a revised unifying model of Rgg regulatory interplay. In contrast to existing models, where Rgg2 proteins are transcriptional activators and Rgg3 proteins are transcriptional repressors, we propose that both are capable of transcriptional activation. However, when Rgg proteins with different activation requirements compete for the same DNA promoters, those with more stringent activation requirements function as repressors by blocking promoter access of SHP-bound conformationally active Rgg proteins. While a similar gene expression regulatory scenario has not been previously described, in all likelihood it is not unique to streptococci. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22341.map.gz emd_22341.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22341-v30.xml emd-22341-v30.xml emd-22341.xml emd-22341.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22341_fsc.xml emd_22341_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_22341.png emd_22341.png | 80.9 KB | ||
| Masks |  emd_22341_msk_1.map emd_22341_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22341.cif.gz emd-22341.cif.gz | 6.4 KB | ||
| Others |  emd_22341_additional_1.map.gz emd_22341_additional_1.map.gz emd_22341_half_map_1.map.gz emd_22341_half_map_1.map.gz emd_22341_half_map_2.map.gz emd_22341_half_map_2.map.gz | 32.2 MB 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22341 http://ftp.pdbj.org/pub/emdb/structures/EMD-22341 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22341 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22341 | HTTPS FTP |
-Related structure data
| Related structure data |  7ji0MC  6w1aC  6w1eC  6w1fC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10535 (Title: Single particle cryoEM of Streptococcus thermophilus SHP pheromone receptor Rgg3 in complex with SHP3 EMPIAR-10535 (Title: Single particle cryoEM of Streptococcus thermophilus SHP pheromone receptor Rgg3 in complex with SHP3Data size: 3.0 TB Data #1: Unaligned multi-frame micrographs of Rgg3-SHP3 complex [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22341.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22341.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map generated by cryoSPARC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.038 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
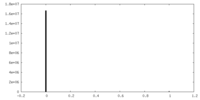
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22341_msk_1.map emd_22341_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
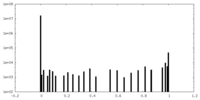
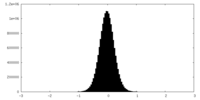
| Density Histograms |
-Additional map: Unsharpened map generated by cryoSPARC
| File | emd_22341_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map generated by cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
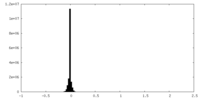
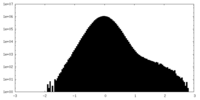
| Density Histograms |
-Half map: Half-map 2 generated by cryoSPARC
| File | emd_22341_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 generated by cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
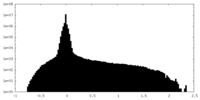
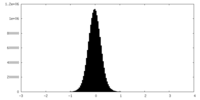
| Density Histograms |
-Half map: Half-map 1 generated by cryoSPARC
| File | emd_22341_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 generated by cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
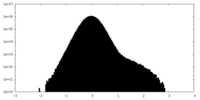
| Density Histograms |
- Sample components
Sample components
-Entire : Streptococcus thermophilus SHP pheromone receptor Rgg3 in complex...
| Entire | Name: Streptococcus thermophilus SHP pheromone receptor Rgg3 in complex with SHP3 |
|---|---|
| Components |
|
-Supramolecule #1: Streptococcus thermophilus SHP pheromone receptor Rgg3 in complex...
| Supramolecule | Name: Streptococcus thermophilus SHP pheromone receptor Rgg3 in complex with SHP3 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus (bacteria) / Organ: CNRZ1066 Streptococcus thermophilus (bacteria) / Organ: CNRZ1066 |
-Macromolecule #1: Positive transcriptional regulator MutR family
| Macromolecule | Name: Positive transcriptional regulator MutR family / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus (strain ATCC BAA-250 / LMG 18311) (bacteria) Streptococcus thermophilus (strain ATCC BAA-250 / LMG 18311) (bacteria)Strain: ATCC BAA-250 / LMG 18311 |
| Molecular weight | Theoretical: 33.242078 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSKLGSTLR KVRNGKQISI CSVADEHLSK SQISRFERGE SEISCIRLIN ILDKLHITLD EFLILHDEDY TKTESFANLV QYIRKQYSL QNINNIQSLL SDSSNYTLDP FEKTMVKSIL HTMDSSIIPS DDELLQLADY LFKVEKWGYY EIILLGNCVR T IDYNSVFL ...String: MKSKLGSTLR KVRNGKQISI CSVADEHLSK SQISRFERGE SEISCIRLIN ILDKLHITLD EFLILHDEDY TKTESFANLV QYIRKQYSL QNINNIQSLL SDSSNYTLDP FEKTMVKSIL HTMDSSIIPS DDELLQLADY LFKVEKWGYY EIILLGNCVR T IDYNSVFL LTKEMLNNYI YSSLNKTNKR IVTQLAINCL ILSIDMEEFT NCFYLIDEIK ALLDNELNFY EQTVFLYATG YF EFKRWQS TSGIEKMKQA IQVLDILGED NLKLHYTIHF DKLINNK UniProtKB: Positive transcriptional regulator MutR family |
-Macromolecule #2: SHP3
| Macromolecule | Name: SHP3 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus thermophilus CNRZ1066 (bacteria) Streptococcus thermophilus CNRZ1066 (bacteria) |
| Molecular weight | Theoretical: 798.969 Da |
| Recombinant expression | Organism: synthetic construct (others) |
| Sequence | String: DIIIIVGG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 3 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV Details: 2.5 microliters Rgg3St-SHP3 was applied to glow-discharged UltraAuFoil (1.2/1.3) 300-mesh grids (Quantifoil), blotted with filter paper for 3-3.5 s, and flash-frozen by plunging in liquid ...Details: 2.5 microliters Rgg3St-SHP3 was applied to glow-discharged UltraAuFoil (1.2/1.3) 300-mesh grids (Quantifoil), blotted with filter paper for 3-3.5 s, and flash-frozen by plunging in liquid ethane cooled with liquid nitrogen. Grids were stored in liquid nitrogen.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 2 / Number real images: 1464 / Average exposure time: 8.0 sec. / Average electron dose: 47.13 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)