+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22239 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Full-length Hsc82 bound to AMPPNP | |||||||||
 Map data Map data | Hsc82 bound to AMPPNP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Hsp90 / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationTetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation / eNOS activation / Extra-nuclear estrogen signaling / VEGFR2 mediated vascular permeability / HSF1-dependent transactivation / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / HSF1 activation / response to oxygen levels / box C/D snoRNP assembly / proteasome assembly ...Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation / eNOS activation / Extra-nuclear estrogen signaling / VEGFR2 mediated vascular permeability / HSF1-dependent transactivation / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / HSF1 activation / response to oxygen levels / box C/D snoRNP assembly / proteasome assembly / Neutrophil degranulation / telomere maintenance / ATP-dependent protein folding chaperone / unfolded protein binding / cellular response to heat / protein folding / protein stabilization / perinuclear region of cytoplasm / ATP hydrolysis activity / protein-containing complex / mitochondrion / ATP binding / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
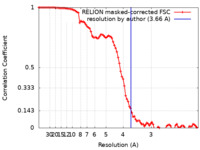
| Method | single particle reconstruction / cryo EM / Resolution: 3.66 Å | |||||||||
 Authors Authors | Liu YX / Sun M | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Full-length Hsc82 bound to AMPPNP Authors: Liu YX / Sun M / Myasnikov AG / Elnatan D / Agard DA | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22239.map.gz emd_22239.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22239-v30.xml emd-22239-v30.xml emd-22239.xml emd-22239.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22239_fsc.xml emd_22239_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_22239.png emd_22239.png | 62.5 KB | ||
| Filedesc metadata |  emd-22239.cif.gz emd-22239.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22239 http://ftp.pdbj.org/pub/emdb/structures/EMD-22239 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22239 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22239 | HTTPS FTP |
-Related structure data
| Related structure data |  6xlcMC  6xlbC  6xldC  6xleC  6xlfC  6xlgC  6xlhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22239.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22239.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hsc82 bound to AMPPNP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
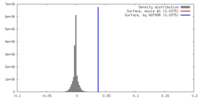
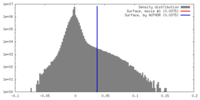
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Hsc82 bound to AMPPNP
| Entire | Name: Hsc82 bound to AMPPNP |
|---|---|
| Components |
|
-Supramolecule #1: Hsc82 bound to AMPPNP
| Supramolecule | Name: Hsc82 bound to AMPPNP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
-Macromolecule #1: ATP-dependent molecular chaperone HSC82
| Macromolecule | Name: ATP-dependent molecular chaperone HSC82 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 81.003594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGETFEFQA EITQLMSLII NTVYSNKEIF LRELISNASD ALDKIRYQAL SDPKQLETEP DLFIRITPKP EEKVLEIRDS GIGMTKAEL INNLGTIAKS GTKAFMEALS AGADVSMIGQ FGVGFYSLFL VADRVQVISK NNEDEQYIWE SNAGGSFTVT L DEVNERIG ...String: MAGETFEFQA EITQLMSLII NTVYSNKEIF LRELISNASD ALDKIRYQAL SDPKQLETEP DLFIRITPKP EEKVLEIRDS GIGMTKAEL INNLGTIAKS GTKAFMEALS AGADVSMIGQ FGVGFYSLFL VADRVQVISK NNEDEQYIWE SNAGGSFTVT L DEVNERIG RGTVLRLFLK DDQLEYLEEK RIKEVIKRHS EFVAYPIQLL VTKEVEKEVP IPEEEKKDEE KKDEDDKKPK LE EVDEEEE EKKPKTKKVK EEVQELEELN KTKPLWTRNP SDITQEEYNA FYKSISNDWE DPLYVKHFSV EGQLEFRAIL FIP KRAPFD LFESKKKKNN IKLYVRRVFI TDEAEDLIPE WLSFVKGVVD SEDLPLNLSR EMLQQNKIMK VIRKNIVKKL IEAF NEIAE DSEQFDKFYS AFAKNIKLGV HEDTQNRAAL AKLLRYNSTK SVDELTSLTD YVTRMPEHQK NIYYITGESL KAVEK SPFL DALKAKNFEV LFLTDPIDEY AFTQLKEFEG KTLVDITKDF ELEETDEEKA EREKEIKEYE PLTKALKDIL GDQVEK VVV SYKLLDAPAA IRTGQFGWSA NMERIMKAQA LRDSSMSSYM SSKKTFEISP KSPIIKELKK RVDEGGAQDK TVKDLTN LL FETALLTSGF SLEEPTSFAS RINRLISLGL NIDEDEETET APEASTEAPV EEVPADTEME EVD UniProtKB: ATP-dependent molecular chaperone HSC82 |
-Macromolecule #2: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 2 / Number of copies: 2 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 72.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6xlc: |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)