+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22113 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Honey bee vitellogenin | |||||||||
 Map data Map data | Vitellogenin (Vg) protein from honey bees | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationnutrient reservoir activity / lipid transporter activity / extracellular region Similarity search - Function | |||||||||
| Biological species |  | |||||||||
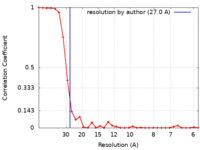
| Method | single particle reconstruction / negative staining / Resolution: 27.0 Å | |||||||||
 Authors Authors | Salmela H / Harwood G / Herrero-Galan E / Amdam G | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Apidologie / Year: 2022 Journal: Apidologie / Year: 2022Title: Nuclear translocation of vitellogenin in the honey bee (). Authors: Heli Salmela / Gyan P Harwood / Daniel Münch / Christine G Elsik / Elías Herrero-Galán / Maria K Vartiainen / Gro V Amdam /     Abstract: Vitellogenin (Vg) is a conserved protein used by nearly all oviparous animals to produce eggs. It is also pleiotropic and performs functions in oxidative stress resistance, immunity, and, in honey ...Vitellogenin (Vg) is a conserved protein used by nearly all oviparous animals to produce eggs. It is also pleiotropic and performs functions in oxidative stress resistance, immunity, and, in honey bees, behavioral development of the worker caste. It has remained enigmatic how Vg affects multiple traits. Here, we asked whether Vg enters the nucleus and acts via DNA-binding. We used cell fractionation, immunohistology, and cell culture to show that a structural subunit of honey bee Vg translocates into cell nuclei. We then demonstrated Vg-DNA binding theoretically and empirically with prediction software and chromatin immunoprecipitation with sequencing (ChIP-seq), finding binding sites at genes influencing immunity and behavior. Finally, we investigated the immunological and enzymatic conditions affecting Vg cleavage and nuclear translocation and constructed a 3D structural model. Our data are the first to show Vg in the nucleus and suggest a new fundamental regulatory role for this ubiquitous protein. SUPPLEMENTARY INFORMATION: The online version contains supplementary material available at 10.1007/s13592-022-00914-9. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22113.map.gz emd_22113.map.gz | 455.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22113-v30.xml emd-22113-v30.xml emd-22113.xml emd-22113.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22113_fsc.xml emd_22113_fsc.xml | 3 KB | Display |  FSC data file FSC data file |
| Images |  emd_22113.png emd_22113.png | 101.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22113 http://ftp.pdbj.org/pub/emdb/structures/EMD-22113 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22113 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22113 | HTTPS FTP |
-Validation report
| Summary document |  emd_22113_validation.pdf.gz emd_22113_validation.pdf.gz | 334.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22113_full_validation.pdf.gz emd_22113_full_validation.pdf.gz | 334.4 KB | Display | |
| Data in XML |  emd_22113_validation.xml.gz emd_22113_validation.xml.gz | 6.3 KB | Display | |
| Data in CIF |  emd_22113_validation.cif.gz emd_22113_validation.cif.gz | 7.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22113 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22113 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22113 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22113 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22113.map.gz / Format: CCP4 / Size: 489.3 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22113.map.gz / Format: CCP4 / Size: 489.3 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Vitellogenin (Vg) protein from honey bees | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.66 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : purified honey bee vitellogenin
| Entire | Name: purified honey bee vitellogenin |
|---|---|
| Components |
|
-Supramolecule #1: purified honey bee vitellogenin
| Supramolecule | Name: purified honey bee vitellogenin / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: vitellogenin
| Macromolecule | Name: vitellogenin / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MLLLLTLLLF AGTVAADFQH NWQVGNEYTY LVRSRTLTSL GDLSDVHTGI LIKALLTVQA KDSNVLAAK VWNGQYARVQ QSMPDGWETE ISDQMLELRD LPISGKPFQI RMKHGLIRDL I VDRDVPTW EVNILKSIVG QLQVDTQGEN AVKVNSVQVP TDDEPYASFK ...String: MLLLLTLLLF AGTVAADFQH NWQVGNEYTY LVRSRTLTSL GDLSDVHTGI LIKALLTVQA KDSNVLAAK VWNGQYARVQ QSMPDGWETE ISDQMLELRD LPISGKPFQI RMKHGLIRDL I VDRDVPTW EVNILKSIVG QLQVDTQGEN AVKVNSVQVP TDDEPYASFK AMEDSVGGKC EV LYDIAPL SDFVIHRSPE LVPMPTLKGD GRHMEVIKIK NFDNCDQRIN YHFGMTDNSR LEP GTNKNG KFFSRSSTSR IVISESLKHF TIQSSVTTSK MMVSPRLYDR QNGLVLSRMN LTLA KMEKT SKPLPMVDNP ESTGNLVYIY NNPFSDVEER RVSKTAMNSN QIVSDNSLSS SEEKL KQDI LNLRTDISSS SSSISSSEEN DFWQPKPTLE DAPQNSLLPN FVGYKGKHIG KSGKVD VIN AAKELIFQIA NELEDASNIP VHATLEKFMI LCNLMRTMNR KQISELESNM QISPNEL KP NDKSQVIKQN TWTVFRDAIT QTGTGPAFLT IKEWIERGTT KSMEAANIMS KLPKTVRT P TDSYIRSFFE LLQNPKVSNE QFLNTAATLS FCEMIHNAQV NKRSIHNNYP VHTFGRLTS KHDNSLYDEY IPFLERELRK AHQEKDSPRI QTYIMALGMI GEPKILSVFE PYLEGKQQMT VFQRTLMVG SLGKLTETNP KLARSVLYKI YLNTMESHEV RCTAVFLLMK TNPPLSMLQR M AEFTKLDT NRQVNSAVKS TIQSLMKLKS PEWKDLAKKA RSVNHLLTHH EYDYELSRGY ID EKILENQ NIITHMILNY VGSEDSVIPR ILYLTWYSSN GDIKVPSTKV LAMISSVKSF MEL SLRSVK DRETIISAAE KIAEELKIVP EELVPLEGNL MINNKYALKF FPFDKHILDK LPTL ISNYI EAVKEGKFMN VNMLDTYESV HSFPTETGLP FVYTFNVIKL TKTSGTVQAQ INPDF AFIV NSNLRLTFSK NVQGRVGFVT PFEHRHFISG IDSNLHVYAP LKISLDVNTP KGNMQW KIW PMKGEEKSRL FHYSVVPFVS NHDILNLRPL SMEKGTRPMI PDDNTSLALP KNEGPFR LN VETAKTNEEM WELIDTEKLT DRLPYPWTMD NERYVKVDMY MNLEGEQKDP VIFSTSFD S KVMTRPDTDS ENWTPKMMAV EPTDKQANSK TRRQEMMREA GRGIESAKSY VVDVRVHVP GESESETVLT LAWSESNVES KGRLLGFWRV EMPRSNADYE VCIGSQIMVS PETLLSYDEK MDQKPKMDF NVDIRYGKNC GKGERIDMNG KLRQSPRLKE LVGATSIIKD CVEDMKRGNK I LRTCQKAV VLSMLLDEVD ISMEVPSDAL IALYSQGLFS LSEIDNLDVS LDVSNPKNAG KK KIDVRAK LNEYLDKADV IVNTPIMDAH FKDVKLSDFG FSTEDILDTA DEDLLINNVF YED ETSCML DKTRAQTFDG KDYPLRLGPC WHAVMTTYPR INPDNHNEKL HIPKDKSVSV LSRE NEAGQ KEVKVLLGSD KIKFVPGTTS QPEVFVNGEK IVVSRNKAYQ KVEENEIIFE IYKMG DRFI GLTSDKFDVS LALDGERVML KASEDYRYSV RGLCGNFDHD STNDFVGPKN CLFRKP EHF VASYALISNQ CEGDSLNVAK SLQDHDCIRQ ERTQQRNVIS DSESGRLDTE MSTWGYH HN VNKHCTIHRT QVKETDDKIC FTMRPVVSCA SGCTAVETKS KPYKFHCMEK NEAAMKLK K RIEKGANPDL SQKPVSTTEE LTVPFVCKA |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Concentration: 150.0 mM / Component - Formula: NaCl / Component - Name: sodium chloride / Details: 20 mM Tris |
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: Aliquotes of pure Vg (1.1 mg/ml in 20 mM Tris, 150 mM NaCl, pH 7.5) were applied to carbon-coated copper grids (30 s) and stained with 2% uranyl acetate |
| Grid | Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1230 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 100 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)