+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-22098 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
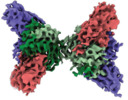
| タイトル | Interleukin-10 signaling complex with IL-10RA and IL-10RB | |||||||||
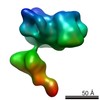
 マップデータ マップデータ | Sharpened map of the hexameric Interleukin-10 signaling complex with IL-10RA and IL-10RB | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | IL-10 / cytokine / receptor / IL-10RA / IL-10RB / signaling | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報interleukin-10 binding / negative regulation of chronic inflammatory response to antigenic stimulus / interleukin-10 receptor binding / regulation of response to wounding / negative regulation of cytokine activity / interleukin-10 receptor activity / negative regulation of interleukin-18 production / interleukin-28 receptor complex / negative regulation of myeloid dendritic cell activation / negative regulation of interferon-alpha production ...interleukin-10 binding / negative regulation of chronic inflammatory response to antigenic stimulus / interleukin-10 receptor binding / regulation of response to wounding / negative regulation of cytokine activity / interleukin-10 receptor activity / negative regulation of interleukin-18 production / interleukin-28 receptor complex / negative regulation of myeloid dendritic cell activation / negative regulation of interferon-alpha production / negative regulation of chemokine (C-C motif) ligand 5 production / positive regulation of cellular respiration / chronic inflammatory response to antigenic stimulus / response to inactivity / negative regulation of membrane protein ectodomain proteolysis / positive regulation of plasma cell differentiation / ubiquitin-dependent endocytosis / positive regulation of B cell apoptotic process / regulation of isotype switching / negative regulation of heterotypic cell-cell adhesion / negative regulation of cytokine production involved in immune response / type III interferon-mediated signaling pathway / negative regulation of MHC class II biosynthetic process / negative regulation of interleukin-1 production / intestinal epithelial structure maintenance / interleukin-10-mediated signaling pathway / branching involved in labyrinthine layer morphogenesis / negative regulation of interleukin-8 production / negative regulation of nitric oxide biosynthetic process / negative regulation of interleukin-12 production / response to carbon monoxide / endothelial cell apoptotic process / positive regulation of MHC class II biosynthetic process / positive regulation of macrophage activation / positive regulation of heterotypic cell-cell adhesion / negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway / leukocyte chemotaxis / negative regulation of mitotic cell cycle / type 2 immune response / CD163 mediating an anti-inflammatory response / Other interleukin signaling / negative regulation of cytokine production / positive regulation of immunoglobulin production / Interleukin-20 family signaling / cellular response to hepatocyte growth factor stimulus / negative regulation of B cell proliferation / defense response to protozoan / regulation of synapse organization / positive regulation of sprouting angiogenesis / Interleukin-10 signaling / negative regulation of interleukin-6 production / negative regulation of type II interferon production / B cell proliferation / hemopoiesis / negative regulation of vascular associated smooth muscle cell proliferation / negative regulation of tumor necrosis factor production / positive regulation of DNA-binding transcription factor activity / Nuclear events stimulated by ALK signaling in cancer / coreceptor activity / positive regulation of vascular associated smooth muscle cell proliferation / negative regulation of T cell proliferation / positive regulation of cell cycle / positive regulation of endothelial cell proliferation / liver regeneration / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / negative regulation of autophagy / FCGR3A-mediated IL10 synthesis / response to glucocorticoid / positive regulation of cytokine production / B cell differentiation / cytokine activity / response to activity / positive regulation of receptor signaling pathway via JAK-STAT / growth factor activity / cellular response to estradiol stimulus / response to molecule of bacterial origin / response to insulin / cellular response to virus / positive regulation of miRNA transcription / negative regulation of inflammatory response / cytokine-mediated signaling pathway / Signaling by ALK fusions and activated point mutants / signaling receptor activity / cellular response to lipopolysaccharide / regulation of gene expression / Interleukin-4 and Interleukin-13 signaling / response to lipopolysaccharide / defense response to virus / protein dimerization activity / defense response to bacterium / immune response / cilium / apical plasma membrane / response to xenobiotic stimulus / inflammatory response / negative regulation of cell population proliferation / negative regulation of apoptotic process / positive regulation of DNA-templated transcription / signal transduction / positive regulation of transcription by RNA polymerase II 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.5 Å | |||||||||
 データ登録者 データ登録者 | Saxton RA / Tsutsumi N | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Science / 年: 2021 ジャーナル: Science / 年: 2021タイトル: Structure-based decoupling of the pro- and anti-inflammatory functions of interleukin-10. 著者: Robert A Saxton / Naotaka Tsutsumi / Leon L Su / Gita C Abhiraman / Kritika Mohan / Lukas T Henneberg / Nanda G Aduri / Cornelius Gati / K Christopher Garcia /  要旨: Interleukin-10 (IL-10) is an immunoregulatory cytokine with both anti-inflammatory and immunostimulatory properties and is frequently dysregulated in disease. We used a structure-based approach to ...Interleukin-10 (IL-10) is an immunoregulatory cytokine with both anti-inflammatory and immunostimulatory properties and is frequently dysregulated in disease. We used a structure-based approach to deconvolute IL-10 pleiotropy by determining the structure of the IL-10 receptor (IL-10R) complex by cryo-electron microscopy at a resolution of 3.5 angstroms. The hexameric structure shows how IL-10 and IL-10Rα form a composite surface to engage the shared signaling receptor IL-10Rβ, enabling the design of partial agonists. IL-10 variants with a range of IL-10Rβ binding strengths uncovered substantial differences in response thresholds across immune cell populations, providing a means of manipulating IL-10 cell type selectivity. Some variants displayed myeloid-biased activity by suppressing macrophage activation without stimulating inflammatory CD8 T cells, thereby uncoupling the major opposing functions of IL-10. These results provide a mechanistic blueprint for tuning the pleiotropic actions of IL-10. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_22098.map.gz emd_22098.map.gz | 59.5 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-22098-v30.xml emd-22098-v30.xml emd-22098.xml emd-22098.xml | 19.4 KB 19.4 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_22098.png emd_22098.png | 269.4 KB | ||
| Filedesc metadata |  emd-22098.cif.gz emd-22098.cif.gz | 6.2 KB | ||
| その他 |  emd_22098_additional_1.map.gz emd_22098_additional_1.map.gz | 32.1 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22098 http://ftp.pdbj.org/pub/emdb/structures/EMD-22098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22098 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_22098_validation.pdf.gz emd_22098_validation.pdf.gz | 507 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_22098_full_validation.pdf.gz emd_22098_full_validation.pdf.gz | 506.6 KB | 表示 | |
| XML形式データ |  emd_22098_validation.xml.gz emd_22098_validation.xml.gz | 6 KB | 表示 | |
| CIF形式データ |  emd_22098_validation.cif.gz emd_22098_validation.cif.gz | 6.9 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22098 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22098 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22098 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22098 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  6x93MC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | |
| 電子顕微鏡画像生データ |  EMPIAR-10557 (タイトル: Interleukin-10 signaling complex with IL-10RA and IL-10RB EMPIAR-10557 (タイトル: Interleukin-10 signaling complex with IL-10RA and IL-10RBData size: 5.4 TB Data #1: Unaligned dark-subtracted TIFF movies with a gain reference for the 3D reconstruction of EMD-22098. [micrographs - multiframe]) |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_22098.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_22098.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Sharpened map of the hexameric Interleukin-10 signaling complex with IL-10RA and IL-10RB | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.078 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-追加マップ: Unsharpened map of the hexameric Interleukin-10 signaling complex...
| ファイル | emd_22098_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Unsharpened map of the hexameric Interleukin-10 signaling complex with IL-10RA and IL-10RB | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
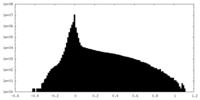
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Interleukin-10 signaling complex with IL-10RA and IL-10RB
| 全体 | 名称: Interleukin-10 signaling complex with IL-10RA and IL-10RB |
|---|---|
| 要素 |
|
-超分子 #1: Interleukin-10 signaling complex with IL-10RA and IL-10RB
| 超分子 | 名称: Interleukin-10 signaling complex with IL-10RA and IL-10RB タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 分子量 | 理論値: 130 KDa |
-超分子 #2: Interleukin-10, IL-10RA
| 超分子 | 名称: Interleukin-10, IL-10RA / タイプ: complex / ID: 2 / 親要素: 1 / 含まれる分子: #1-#2 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-超分子 #3: IL-10RB
| 超分子 | 名称: IL-10RB / タイプ: complex / ID: 3 / 親要素: 1 / 含まれる分子: #3 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Interleukin-10
| 分子 | 名称: Interleukin-10 / タイプ: protein_or_peptide / ID: 1 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 18.778543 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: SPGQGTQSEN SCTHFPGYLP NMLRDLRDAF SRVKTFFQMK DQLDNLLLKE SLLEDFKGYL GCQALSEMIQ FYLEEVMPQA ENQDPDIKA HVQSLGENLK DLRLWLRRCH RFLPCENKSK AVEQVKNAFN KLQEKGIYKA MSEFDIFINY IEAYMTMKIR N UniProtKB: Interleukin-10 |
-分子 #2: Interleukin-10 receptor subunit alpha
| 分子 | 名称: Interleukin-10 receptor subunit alpha / タイプ: protein_or_peptide / ID: 2 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 24.422391 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: HGTELPSPPS VWFEAEFFHH ILHWTPIPNQ SESTCYEVAL LRYGIESWNS ISNCSQTLSY DLTAVTLDLY HSNGYRARVR AVDGSRHSN WTVTNTRFSV DEVTLTVGSV NLEIHNGFIL GKIQLPRPKM APANDTYESI FSHFREYEIA IRKVPGNFTF T HKKVKHEN ...文字列: HGTELPSPPS VWFEAEFFHH ILHWTPIPNQ SESTCYEVAL LRYGIESWNS ISNCSQTLSY DLTAVTLDLY HSNGYRARVR AVDGSRHSN WTVTNTRFSV DEVTLTVGSV NLEIHNGFIL GKIQLPRPKM APANDTYESI FSHFREYEIA IRKVPGNFTF T HKKVKHEN FSLLTSGEVG EFCVQVKPSV ASRSNKGMWS KEECISLTRQ YFTVTN UniProtKB: Interleukin-10 receptor subunit alpha |
-分子 #3: Interleukin-10 receptor subunit beta
| 分子 | 名称: Interleukin-10 receptor subunit beta / タイプ: protein_or_peptide / ID: 3 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 23.569334 KDa |
| 組換発現 | 生物種:  Trichoplusia ni (イラクサキンウワバ) Trichoplusia ni (イラクサキンウワバ) |
| 配列 | 文字列: MVPPPENVRM NSVNFKNILQ WESPAFAKGQ LTFTAQYLSY RIFQDKCMQT TLTECDFSSL SKYGDHTLRV RAEFADEHSD WVQITFCPV DDTIIGPPGM QVEVLADSLH MRFLAPKIEN EYETWTMKNV YNSWTYNVQY WKNGTDEKFQ ITPQYDFEVL R NLEPWTTY ...文字列: MVPPPENVRM NSVNFKNILQ WESPAFAKGQ LTFTAQYLSY RIFQDKCMQT TLTECDFSSL SKYGDHTLRV RAEFADEHSD WVQITFCPV DDTIIGPPGM QVEVLADSLH MRFLAPKIEN EYETWTMKNV YNSWTYNVQY WKNGTDEKFQ ITPQYDFEVL R NLEPWTTY CVQVRGFLPD RNKAGEWSEP VCEQTTHDET VPS UniProtKB: Interleukin-10 receptor subunit beta |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 10 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.2 構成要素:
| ||||||||||||
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: GOLD / メッシュ: 300 / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 40 sec. | ||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 95 % / チャンバー内温度: 293 K / 装置: LEICA EM GP / 詳細: 5s blotting. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 特殊光学系 | エネルギーフィルター - 名称: GIF Bioquantum / エネルギーフィルター - スリット幅: 20 eV |
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 撮影したグリッド数: 1 / 実像数: 9413 / 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 100.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): -2.0 µm / 最小 デフォーカス(公称値): -0.8 µm / 倍率(公称値): 81000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)