[English] 日本語
 Yorodumi
Yorodumi- EMDB-22067: Bovine Cardiac Myosin in Complex with Chicken Skeletal Actin and ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22067 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bovine Cardiac Myosin in Complex with Chicken Skeletal Actin and Human Cardiac Tropomyosin in the Rigor State | ||||||||||||||||||||||||
 Map data Map data | Cryo-EM map of the cardiac actin-myosin-tropomyosin complex. | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | myosin / tropomyosin / actin / cardiac / CONTRACTILE PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of heart rate by epinephrine / muscle thin filament tropomyosin / Striated Muscle Contraction / regulation of muscle contraction / bleb / myosin filament / ruffle organization / adult heart development / Striated Muscle Contraction / muscle filament sliding ...positive regulation of heart rate by epinephrine / muscle thin filament tropomyosin / Striated Muscle Contraction / regulation of muscle contraction / bleb / myosin filament / ruffle organization / adult heart development / Striated Muscle Contraction / muscle filament sliding / myosin II complex / sarcomere organization / structural constituent of muscle / ventricular cardiac muscle tissue morphogenesis / microfilament motor activity / negative regulation of vascular associated smooth muscle cell migration / regulation of heart contraction / myofibril / negative regulation of vascular associated smooth muscle cell proliferation / striated muscle thin filament / skeletal muscle thin filament assembly / Smooth Muscle Contraction / skeletal muscle fiber development / cardiac muscle contraction / stress fiber / cytoskeletal protein binding / positive regulation of stress fiber assembly / cytoskeleton organization / positive regulation of cell adhesion / negative regulation of cell migration / actin filament organization / sarcomere / actin filament / cellular response to reactive oxygen species / wound healing / structural constituent of cytoskeleton / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / ruffle membrane / actin filament binding / regulation of cell shape / actin cytoskeleton / actin binding / cytoskeleton / calmodulin binding / protein heterodimerization activity / hydrolase activity / protein homodimerization activity / ATP binding / identical protein binding / cytoplasm / cytosol Similarity search - Function | ||||||||||||||||||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||||||||
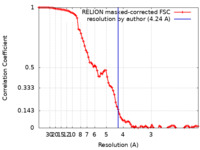
| Method | helical reconstruction / cryo EM / Resolution: 4.24 Å | ||||||||||||||||||||||||
 Authors Authors | Doran MH / Lehman W / Bullitt E | ||||||||||||||||||||||||
| Funding support |  United States, European Union, 7 items United States, European Union, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Biophys J / Year: 2020 Journal: Biophys J / Year: 2020Title: Cryo-EM and Molecular Docking Shows Myosin Loop 4 Contacts Actin and Tropomyosin on Thin Filaments. Authors: Matthew H Doran / Elumalai Pavadai / Michael J Rynkiewicz / Jonathan Walklate / Esther Bullitt / Jeffrey R Moore / Michael Regnier / Michael A Geeves / William Lehman /   Abstract: The motor protein myosin drives muscle and nonmuscle motility by binding to and moving along actin of thin filaments. Myosin binding to actin also modulates interactions of the regulatory protein, ...The motor protein myosin drives muscle and nonmuscle motility by binding to and moving along actin of thin filaments. Myosin binding to actin also modulates interactions of the regulatory protein, tropomyosin, on thin filaments, and conversely tropomyosin affects myosin binding to actin. Insight into this reciprocity will facilitate a molecular level elucidation of tropomyosin regulation of myosin interaction with actin in muscle contraction, and in turn, promote better understanding of nonmuscle cell motility. Indeed, experimental approaches such as fiber diffraction, cryoelectron microscopy, and three-dimensional reconstruction have long been used to define regulatory interaction of tropomyosin and myosin on actin at a structural level. However, their limited resolution has not proven sufficient to determine tropomyosin and myosin contacts at an atomic-level and thus to fully substantiate possible functional contributions. To overcome this deficiency, we have followed a hybrid approach by performing new cryogenic electron microscopy reconstruction of myosin-S1-decorated F-actin-tropomyosin together with atomic scale protein-protein docking of tropomyosin to the EM models. Here, cryo-EM data were derived from filaments reconstituted with α1-actin, cardiac αα-tropomyosin, and masseter muscle β-myosin complexes; masseter myosin, which shares sequence identity with β-cardiac myosin-heavy chain, was used because of its stability in vitro. The data were used to build an atomic model of the tropomyosin cable that fits onto the actin filament between the tip of the myosin head and a cleft on the innermost edge of actin subunits. The docking and atomic scale fitting showed multiple discrete interactions of myosin loop 4 and acidic residues on successive 39-42 residue-long tropomyosin pseudorepeats. The contacts between S1 and tropomyosin on actin appear to compete with and displace ones normally found between actin and tropomyosin on myosin-free thin filaments in relaxed muscle, thus restructuring the filament during myosin-induced activation. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22067.map.gz emd_22067.map.gz | 9.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22067-v30.xml emd-22067-v30.xml emd-22067.xml emd-22067.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22067_fsc.xml emd_22067_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_22067.png emd_22067.png | 222.1 KB | ||
| Filedesc metadata |  emd-22067.cif.gz emd-22067.cif.gz | 8.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22067 http://ftp.pdbj.org/pub/emdb/structures/EMD-22067 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22067 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22067 | HTTPS FTP |
-Related structure data
| Related structure data |  6x5zMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22067.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22067.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of the cardiac actin-myosin-tropomyosin complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Helical complex of bovine S1 cardiac myosin and human cardiac tro...
| Entire | Name: Helical complex of bovine S1 cardiac myosin and human cardiac tropomyosin-decorated chicken skeletal actin filaments |
|---|---|
| Components |
|
-Supramolecule #1: Helical complex of bovine S1 cardiac myosin and human cardiac tro...
| Supramolecule | Name: Helical complex of bovine S1 cardiac myosin and human cardiac tropomyosin-decorated chicken skeletal actin filaments type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Bovine myosin was isolated from the masseter muscle and the S1 fragment was generated through proteolytic cleavage. |
|---|---|
| Molecular weight | Theoretical: 403.22 KDa |
-Supramolecule #3: S1 cardiac myosin
| Supramolecule | Name: S1 cardiac myosin / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #4: cardiac tropomyosin
| Supramolecule | Name: cardiac tropomyosin / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: skeletal actin filaments
| Supramolecule | Name: skeletal actin filaments / type: organelle_or_cellular_component / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 1 Details: Residues 1-9 and 377 were disordered and were not modeled. Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.096953 KDa |
| Sequence | String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIEHGIITN WDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLDSGD G VTHNVPIY ...String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIEHGIITN WDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLDSGD G VTHNVPIY EGYALPHAIM RLDLAGRDLT DYLMKILTER GYSFVTTAER EIVRDIKEKL CYVALDFENE MATAASSSSL EK SYELPDG QVITIGNERF RCPETLFQPS FIGMESAGIH ETTYNSIMKC DIDIRKDLYA NNVMSGGTTM YPGIADRMQK EIT ALAPST MKIKIIAPPE RKYSVWIGGS ILASLSTFQQ MWITKQEYDE AGPSIVHRKC F UniProtKB: Actin, alpha skeletal muscle |
-Macromolecule #2: Tropomyosin alpha-1 chain
| Macromolecule | Name: Tropomyosin alpha-1 chain / type: protein_or_peptide / ID: 2 Details: Only the central section (residues 45-210) was modeled. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.763621 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDAIKKKMQM LKLDKENALD RAEQAEADKK AAEDRSKQLE DELVSLQKKL KGTEDELDKY SEALKDAQEK LELAEKKATD AEADVASLN RRIQLVEEEL DRAQERLATA LQKLEEAEKA ADESERGMKV IESRAQKDEE KMEIQEIQLK EAKHIAEDAD R KYEEVARK ...String: MDAIKKKMQM LKLDKENALD RAEQAEADKK AAEDRSKQLE DELVSLQKKL KGTEDELDKY SEALKDAQEK LELAEKKATD AEADVASLN RRIQLVEEEL DRAQERLATA LQKLEEAEKA ADESERGMKV IESRAQKDEE KMEIQEIQLK EAKHIAEDAD R KYEEVARK LVIIESDLER AEERAELSEG KCAELEEELK TVTNNLKSLE AQAEKYSQKE DRYEEEIKVL SDKLKEAETR AE FAERSVT KLEKSIDDLE DELYAQKLKY KAISEELDHA LNDMTSI UniProtKB: Tropomyosin alpha-1 chain |
-Macromolecule #3: Myosin-7
| Macromolecule | Name: Myosin-7 / type: protein_or_peptide / ID: 3 Details: Residues 1-35, 200-216, 624-641, and 777-850 were disordered and not modeled. Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 97.446898 KDa |
| Sequence | String: MVDAEMAAFG EAAPYLRKSE KERLEAQTRP FDLKKDVFVP DDKEEFVKAT ILSREGGKVT AETEHGKTVT VKEDQVLQQN PPKFDKIED MAMLTFLHEP AVLYNLKERY ASWMIYTYSG LFCVTINPYK WLPVYNAEVV AAYRGKKRSE APPHIFSISD N AYQYMLTD ...String: MVDAEMAAFG EAAPYLRKSE KERLEAQTRP FDLKKDVFVP DDKEEFVKAT ILSREGGKVT AETEHGKTVT VKEDQVLQQN PPKFDKIED MAMLTFLHEP AVLYNLKERY ASWMIYTYSG LFCVTINPYK WLPVYNAEVV AAYRGKKRSE APPHIFSISD N AYQYMLTD RENQSILITG ESGAGKTVNT KRVIQYFAVI AAIGDRSKKE QATGKGTLED QIIQANPALE AFGNAKTVRN DN SSRFGKF IRIHFGATGK LASADIETYL LEKSRVIFQL KAERDYHIFY QILSNKKPEL LDMLLITNNP YDYAFISQGE TTV ASIDDA EELMATDNAF DVLGFTTEEK NSMYKLTGAI MHFGNMKFKL KQREEQAEPD GTEEADKSAY LMGLNSADLL KGLC HPRVK VGNEYVTKGQ NVQQVVYAKG ALAKAVYERM FNWMVTRINA TLETKQPRQY FIGVLDIAGF EIFDFNSFEQ LCINF TNEK LQQFFNHHMF VLEQEEYKKE GIEWEFIDFG MDLQACIDLI EKPMGIMSIL EEECMFPKAT DMTFKAKLFD NHLGKS SNF QKPRNIKGKP EAHFSLIHYA GTVDYNIIGW LQKNKDPLNE TVVDLYKKSS LKMLSSLFAN YAGFDTPIEK GKGKAKK GS SFQTVSALHR ENLNKLMTNL RSTHPHFVRC IIPNETKSPG VIDNPLVMHQ LRCNGVLEGI RICRKGFPNR ILYGDFRQ R YRILNPAAIP EGQFIDSRKG AEKLLGSLDI DHNQYKFGHT KVFFKAGLLG LLEEMRDERL SRIITRIQAQ SRGVLSRME FKKLLERRDS LLIIQWNIRA FMGVKNWPWM KLYFKIKPLL KSAETEKEIA UniProtKB: Myosin-7 |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 3 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | .525 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: AIR / Details: Glow discharged using a PELCO easiGlow station | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK III Details: Thin filaments were reconstituted by first mixing F-actin and tropomyosin to final concentrations of 10 uM actin and 7 uM tropomyosin. Just before applying 1.5 uL actin-tropomyosin to a ...Details: Thin filaments were reconstituted by first mixing F-actin and tropomyosin to final concentrations of 10 uM actin and 7 uM tropomyosin. Just before applying 1.5 uL actin-tropomyosin to a freshly glow discharged holey-carbon grid, the surfactant octyl B-D-glucopyranoside was added to the protein solution to a concentration of 12 nM. The grid sample was manually blotted for 1 second and a 1.5 uL drop of 7.5 uM myosin-S1 sub-fragment was then applied to the blotted grid sample. The sample was immediately blotted for 4 seconds and plunge-frozen in liquid ethane.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-35 / Number grids imaged: 1 / Number real images: 2496 / Average exposure time: 7.0 sec. / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 130000 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Details | version 1.9.3; initial flexible fitting to align secondary structure using MDFF | |||||||||
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | |||||||||
| Output model |  PDB-6x5z: |
-Atomic model buiding 2
| Details | version 1.18-3861 |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 70.88 |
| Output model |  PDB-6x5z: |
-Atomic model buiding 3
| Details | manual refinement of residues to match backbone and sidechain density and to limit clashes, sidechain outliers, and Ramachandran outliers |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER |
| Output model |  PDB-6x5z: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)