+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22042 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MCU-EMRE complex of a metazoan mitochondrial calcium uniporter | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channel / calcium channel / mitochondrial calcium uniporter / MCU / EMRE / mitochondria / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationuniporter activity / uniplex complex / mitochondrial calcium ion homeostasis / calcium import into the mitochondrion / calcium channel activity / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Long SB / Wang C | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2020 Journal: J Mol Biol / Year: 2020Title: Structure and Reconstitution of an MCU-EMRE Mitochondrial Ca Uniporter Complex. Authors: Chongyuan Wang / Rozbeh Baradaran / Stephen Barstow Long /  Abstract: The proteins MCU and EMRE form the minimal functional unit of the mitochondrial calcium uniporter complex in metazoans, a highly selective and tightly controlled Ca channel of the inner mitochondrial ...The proteins MCU and EMRE form the minimal functional unit of the mitochondrial calcium uniporter complex in metazoans, a highly selective and tightly controlled Ca channel of the inner mitochondrial membrane that regulates cellular metabolism. Here we present functional reconstitution of an MCU-EMRE complex from the red flour beetle, Tribolium castaneum, and a cryo-EM structure of the complex at 3.5 Å resolution. Using a novel assay, we demonstrate robust Ca uptake into proteoliposomes containing the purified complex. Uptake is dependent on EMRE and also on the mitochondrial lipid cardiolipin. The structure reveals a tetrameric channel with a single ion pore. EMRE is located at the periphery of the transmembrane domain and associates primarily with the first transmembrane helix of MCU. Coiled-coil and juxtamembrane domains within the matrix portion of the complex adopt markedly different conformations than in a structure of a human MCU-EMRE complex, suggesting that the structures represent different conformations of these functionally similar metazoan channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22042.map.gz emd_22042.map.gz | 1.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22042-v30.xml emd-22042-v30.xml emd-22042.xml emd-22042.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22042.png emd_22042.png | 113 KB | ||
| Filedesc metadata |  emd-22042.cif.gz emd-22042.cif.gz | 6.1 KB | ||
| Others |  emd_22042_additional.map.gz emd_22042_additional.map.gz | 95.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22042 http://ftp.pdbj.org/pub/emdb/structures/EMD-22042 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22042 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22042 | HTTPS FTP |
-Validation report
| Summary document |  emd_22042_validation.pdf.gz emd_22042_validation.pdf.gz | 325.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22042_full_validation.pdf.gz emd_22042_full_validation.pdf.gz | 324.6 KB | Display | |
| Data in XML |  emd_22042_validation.xml.gz emd_22042_validation.xml.gz | 6.4 KB | Display | |
| Data in CIF |  emd_22042_validation.cif.gz emd_22042_validation.cif.gz | 7.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22042 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22042 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22042 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22042 | HTTPS FTP |
-Related structure data
| Related structure data |  6x4sMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22042.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22042.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.088 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
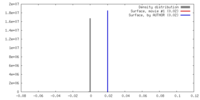
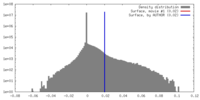
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unsharpened map
| File | emd_22042_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
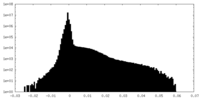
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mitochondrial Calcium Uniporter (MCU) in complex with EMRE. Embed...
| Entire | Name: Mitochondrial Calcium Uniporter (MCU) in complex with EMRE. Embedded in lipid nanodiscs containing cardiolipin. |
|---|---|
| Components |
|
-Supramolecule #1: Mitochondrial Calcium Uniporter (MCU) in complex with EMRE. Embed...
| Supramolecule | Name: Mitochondrial Calcium Uniporter (MCU) in complex with EMRE. Embedded in lipid nanodiscs containing cardiolipin. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Calcium uniporter protein,Protein EMRE homolog, mitochondrial-lik...
| Macromolecule | Name: Calcium uniporter protein,Protein EMRE homolog, mitochondrial-like Protein fusion type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.530723 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPTAAALERL TTEEVQGLSD VKTLVNQLYE ALNVREHQLQ KEVELTTQLE TLQQELLPLE EKKLELEQVA NRRSNWMAWA GLGLMSVQF GILARLTWWE YSWDIMEPVT YFVTYGTAMA AYAYFVLTRE EYILNDVRDR QQLLLLHKKA KKTGFDVNQY N VLKDQIAK ...String: GPTAAALERL TTEEVQGLSD VKTLVNQLYE ALNVREHQLQ KEVELTTQLE TLQQELLPLE EKKLELEQVA NRRSNWMAWA GLGLMSVQF GILARLTWWE YSWDIMEPVT YFVTYGTAMA AYAYFVLTRE EYILNDVRDR QQLLLLHKKA KKTGFDVNQY N VLKDQIAK LELDLKRLRD PLKLRLPPKA AASGSGSGEN LYFQGSGGLL PEPHRTSFGI IRLILTVVPG LLIGAAISKN IA NFL UniProtKB: Calcium uniporter protein, mitochondrial, Essential MCU regulator, mitochondrial |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: 2 second blot, blot force of 0. | ||||||||||||
| Details | Monodisperse sample |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Number grids imaged: 3 / Number real images: 8143 / Average exposure time: 0.25 sec. / Average electron dose: 1.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: -3.0 µm / Nominal defocus min: -1.0 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 127 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-6x4s: |
 Movie
Movie Controller
Controller



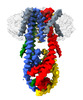








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)