[English] 日本語
 Yorodumi
Yorodumi- EMDB-21389: Negative Staining Reconstruction of Tryptase Tetramer bound to E1... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21389 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
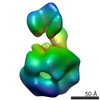
| Title | Negative Staining Reconstruction of Tryptase Tetramer bound to E104 Fab | |||||||||
 Map data Map data | Tryptase bound to E104 Fab | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 15.0 Å | |||||||||
 Authors Authors | Koerber JT / Lazarus B / Maun H / Estevez A / Ciferri C | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Bivalent antibody pliers inhibit β-tryptase by an allosteric mechanism dependent on the IgG hinge. Authors: Henry R Maun / Rajesh Vij / Benjamin T Walters / Ashley Morando / Janet K Jackman / Ping Wu / Alberto Estevez / Xiaocheng Chen / Yvonne Franke / Michael T Lipari / Mark S Dennis / Daniel ...Authors: Henry R Maun / Rajesh Vij / Benjamin T Walters / Ashley Morando / Janet K Jackman / Ping Wu / Alberto Estevez / Xiaocheng Chen / Yvonne Franke / Michael T Lipari / Mark S Dennis / Daniel Kirchhofer / Claudio Ciferri / Kelly M Loyet / Tangsheng Yi / Charles Eigenbrot / Robert A Lazarus / James T Koerber /  Abstract: Human β-tryptase, a tetrameric trypsin-like serine protease, is an important mediator of allergic inflammatory responses in asthma. Antibodies generally inhibit proteases by blocking substrate ...Human β-tryptase, a tetrameric trypsin-like serine protease, is an important mediator of allergic inflammatory responses in asthma. Antibodies generally inhibit proteases by blocking substrate access by binding to active sites or exosites or by allosteric modulation. The bivalency of IgG antibodies can increase potency via avidity, but has never been described as essential for activity. Here we report an inhibitory anti-tryptase IgG antibody with a bivalency-driven mechanism of action. Using biochemical and structural data, we determine that four Fabs simultaneously occupy four exosites on the β-tryptase tetramer, inducing allosteric changes at the small interface. In the presence of heparin, the monovalent Fab shows essentially no inhibition, whereas the bivalent IgG fully inhibits β-tryptase activity in a hinge-dependent manner. Our results suggest a model where the bivalent IgG acts akin to molecular pliers, pulling the tetramer apart into inactive β-tryptase monomers, and may provide an alternative strategy for antibody engineering. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21389.map.gz emd_21389.map.gz | 22.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21389-v30.xml emd-21389-v30.xml emd-21389.xml emd-21389.xml | 9.2 KB 9.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21389.png emd_21389.png | 172.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21389 http://ftp.pdbj.org/pub/emdb/structures/EMD-21389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21389 | HTTPS FTP |
-Validation report
| Summary document |  emd_21389_validation.pdf.gz emd_21389_validation.pdf.gz | 301 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21389_full_validation.pdf.gz emd_21389_full_validation.pdf.gz | 300.6 KB | Display | |
| Data in XML |  emd_21389_validation.xml.gz emd_21389_validation.xml.gz | 5.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21389 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21389 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21389 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21389 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21389.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21389.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tryptase bound to E104 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.73 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Tryptase bound to E104 Fab
| Entire | Name: Tryptase bound to E104 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Tryptase bound to E104 Fab
| Supramolecule | Name: Tryptase bound to E104 Fab / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 340 kDa/nm |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 6.8 / Details: 10 mM MOPS pH 6.8, 2 M NaCl |
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: Four microliters of purified beta-tryptase bound to E104.v1 Fab fragment were applied to a freshly glow discharged 400-mesh copper EM grid covered with a thin layer of continuous carbon (Ted ...Details: Four microliters of purified beta-tryptase bound to E104.v1 Fab fragment were applied to a freshly glow discharged 400-mesh copper EM grid covered with a thin layer of continuous carbon (Ted Pella, Redding, CA). After 30 s of incubation, the grids were stained with 5 drops of a freshly prepared 0.75% (w/v) uranyl formate solution. |
| Grid | Pretreatment - Type: GLOW DISCHARGE / Details: unspecified |
| Details | purified through affinity purification and Size Exclusion Chromatography in 10 mM MOPS pH 6.8, 2 M NaCl. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 15.0 Å / Resolution method: OTHER / Number images used: 15000 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















