[English] 日本語
 Yorodumi
Yorodumi- EMDB-21232: BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20E1 and PG... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21232 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20E1 and PGT122 Fab | ||||||||||||||||||
 Map data Map data | BG505 SOSIP.v5.2 in complex with Rhesus macaque Fab RM20E1 and PGT122 Fab | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | HIV / antibody / Fab / vaccine / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | ||||||||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||
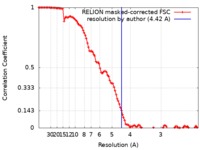
| Method | single particle reconstruction / cryo EM / Resolution: 4.42 Å | ||||||||||||||||||
 Authors Authors | Cottrell CA / Ward AB / Patel R | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2020 Journal: PLoS Pathog / Year: 2020Title: Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates. Authors: Christopher A Cottrell / Jelle van Schooten / Charles A Bowman / Meng Yuan / David Oyen / Mia Shin / Robert Morpurgo / Patricia van der Woude / Mariëlle van Breemen / Jonathan L Torres / ...Authors: Christopher A Cottrell / Jelle van Schooten / Charles A Bowman / Meng Yuan / David Oyen / Mia Shin / Robert Morpurgo / Patricia van der Woude / Mariëlle van Breemen / Jonathan L Torres / Raj Patel / Justin Gross / Leigh M Sewall / Jeffrey Copps / Gabriel Ozorowski / Bartek Nogal / Devin Sok / Eva G Rakasz / Celia Labranche / Vladimir Vigdorovich / Scott Christley / Diane G Carnathan / D Noah Sather / David Montefiori / Guido Silvestri / Dennis R Burton / John P Moore / Ian A Wilson / Rogier W Sanders / Andrew B Ward / Marit J van Gils /   Abstract: The induction of broad and potent immunity by vaccines is the key focus of research efforts aimed at protecting against HIV-1 infection. Soluble native-like HIV-1 envelope glycoproteins have shown ...The induction of broad and potent immunity by vaccines is the key focus of research efforts aimed at protecting against HIV-1 infection. Soluble native-like HIV-1 envelope glycoproteins have shown promise as vaccine candidates as they can induce potent autologous neutralizing responses in rabbits and non-human primates. In this study, monoclonal antibodies were isolated and characterized from rhesus macaques immunized with the BG505 SOSIP.664 trimer to better understand vaccine-induced antibody responses. Our studies reveal a diverse landscape of antibodies recognizing immunodominant strain-specific epitopes and non-neutralizing neo-epitopes. Additionally, we isolated a subset of mAbs against an epitope cluster at the gp120-gp41 interface that recognize the highly conserved fusion peptide and the glycan at position 88 and have characteristics akin to several human-derived broadly neutralizing antibodies. #1:  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: Mapping the immunogenic landscape of native HIV-1 envelope trimers in non-human primates Authors: Cottrell CA / van Schooten J / Bowman CA / Yuan M / Oyen D / Shin M / Morpurgo R / van der Woude P / van Breemen M / Torres JL / Patel R / Gross J / Sewall LM / Copps J / Ozorowski G / Sok D ...Authors: Cottrell CA / van Schooten J / Bowman CA / Yuan M / Oyen D / Shin M / Morpurgo R / van der Woude P / van Breemen M / Torres JL / Patel R / Gross J / Sewall LM / Copps J / Ozorowski G / Sok D / Rakasz EG / Labranche C / Vigdorovich V / Christley S / Sather DN / Montefiori D / Moore JP / Burton DR / Wilson IA / Sanders RW / Ward AB / van Gils MJ | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21232.map.gz emd_21232.map.gz | 95.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21232-v30.xml emd-21232-v30.xml emd-21232.xml emd-21232.xml | 25.8 KB 25.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21232_fsc.xml emd_21232_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_21232.png emd_21232.png | 131.6 KB | ||
| Masks |  emd_21232_msk_1.map emd_21232_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-21232.cif.gz emd-21232.cif.gz | 7.4 KB | ||
| Others |  emd_21232_half_map_1.map.gz emd_21232_half_map_1.map.gz emd_21232_half_map_2.map.gz emd_21232_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21232 http://ftp.pdbj.org/pub/emdb/structures/EMD-21232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21232 | HTTPS FTP |
-Validation report
| Summary document |  emd_21232_validation.pdf.gz emd_21232_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21232_full_validation.pdf.gz emd_21232_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_21232_validation.xml.gz emd_21232_validation.xml.gz | 17.6 KB | Display | |
| Data in CIF |  emd_21232_validation.cif.gz emd_21232_validation.cif.gz | 23.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21232 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21232 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21232 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21232 | HTTPS FTP |
-Related structure data
| Related structure data |  6vlrMC  6vn0C  6vo1C  6vorC  6vosC  6vsrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21232.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21232.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
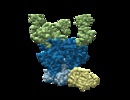
| Annotation | BG505 SOSIP.v5.2 in complex with Rhesus macaque Fab RM20E1 and PGT122 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
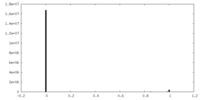
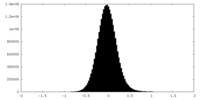
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21232_msk_1.map emd_21232_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
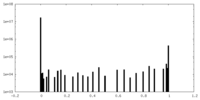
| Projections & Slices |
| ||||||||||||
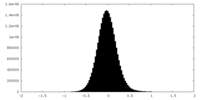
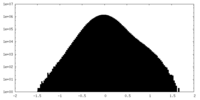
| Density Histograms |
-Half map: BG505 SOSIP.v5.2 in complex with Rhesus macaque Fab...
| File | emd_21232_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BG505 SOSIP.v5.2 in complex with Rhesus macaque Fab RM20E1 and PGT122 Fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
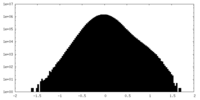
| Density Histograms |
-Half map: BG505 SOSIP.v5.2 in complex with Rhesus macaque Fab...
| File | emd_21232_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BG505 SOSIP.v5.2 in complex with Rhesus macaque Fab RM20E1 and PGT122 Fab | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20E1
| Entire | Name: BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20E1 |
|---|---|
| Components |
|
-Supramolecule #1: BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20E1
| Supramolecule | Name: BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20E1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 53.268371 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETKKHNVWA THCCVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETKKHNVWA THCCVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT SAITQACPKV SFEPIPIHYC APAGFAILKC KDKKFNGTGP CPSVSTVQCT HGIKPVVSTQ LLLNGSLAEE EV MIRSENI TNNAKNILVQ FNTPVQINCT RPNNNTRKSI RIGPGQWFYA TGDIIGDIRQ AHCNVSKATW NETLGKVVKQ LRK HFGNNT IIRFANSSGG DLEVTTHSFN CGGEFFYCNT SGLFNSTWIS NTSVQGSNST GSNDSITLPC RIKQIINMWQ RIGQ AMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRVVG UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: BG505 SOSIPv5.2 gp41
| Macromolecule | Name: BG505 SOSIPv5.2 gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.178549 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEC QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: PGT122 Fab Heavy Chain
| Macromolecule | Name: PGT122 Fab Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.838731 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVHLQESGPG LVKPSETLSL TCNVSGTLVR DNYWSWIRQP LGKQPEWIGY VHDSGDTNYN PSLKSRVHLS LDKSKNLVSL RLTGVTAAD SAIYYCATTK HGRRIYGVVA FKEWFTYFYM DVWGKGTSVT VSS |
-Macromolecule #4: PGT122 Fab Light Chain
| Macromolecule | Name: PGT122 Fab Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.510612 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TFVSVAPGQT ARITCGEESL GSRSVIWYQQ RPGQAPSLII YNNNDRPSGI PDRFSGSPGS TFGTTATLTI TSVEAGDEAD YYCHIWDSR RPTNWVFGEG TTLIVLS |
-Macromolecule #5: RM20E1 Fab Heavy Chain
| Macromolecule | Name: RM20E1 Fab Heavy Chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.878558 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVQSEAE VKRPGESLKI SCQTSGYNFP NYWITWVRQM PGKGLEWMGT IDPRDSDTKY SPSFQGQVTI SADKSINTAY LQWTSLRAS DSATYYCVMW VYILTTGNIW VDVWGPGVLV TVSS |
-Macromolecule #6: RM20E1 Fab Kappa Chain
| Macromolecule | Name: RM20E1 Fab Kappa Chain / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.370759 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DVVMTQSPLS LPITPGQPAS ISCRSSQSLV HNNGNTYLTW YQQRPGQPPR RLIYQVSNRD SGVPDRFIGS GAGTDFTLKI SRVESEDVG IYYCGQITDF PYSFGQGTKV DIK |
-Macromolecule #12: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 12 / Number of copies: 24 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 51.3 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-6vlr: |
 Movie
Movie Controller
Controller













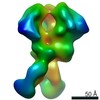



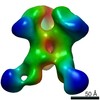
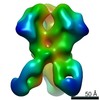

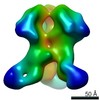


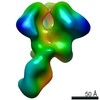

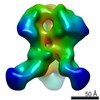


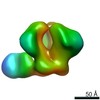





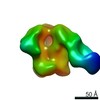















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)