+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21131 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of TRPA1 treated with A-967079 (class 2), PMAL-C8 | |||||||||
 Map data Map data | Full map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Zhao J / Lin King JV / Paulsen CE / Cheng Y / Julius D | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Irritant-evoked activation and calcium modulation of the TRPA1 receptor. Authors: Jianhua Zhao / John V Lin King / Candice E Paulsen / Yifan Cheng / David Julius /  Abstract: The transient receptor potential ion channel TRPA1 is expressed by primary afferent nerve fibres, in which it functions as a low-threshold sensor for structurally diverse electrophilic irritants, ...The transient receptor potential ion channel TRPA1 is expressed by primary afferent nerve fibres, in which it functions as a low-threshold sensor for structurally diverse electrophilic irritants, including small volatile environmental toxicants and endogenous algogenic lipids. TRPA1 is also a 'receptor-operated' channel whose activation downstream of metabotropic receptors elicits inflammatory pain or itch, making it an attractive target for novel analgesic therapies. However, the mechanisms by which TRPA1 recognizes and responds to electrophiles or cytoplasmic second messengers remain unknown. Here we use strutural studies and electrophysiology to show that electrophiles act through a two-step process in which modification of a highly reactive cysteine residue (C621) promotes reorientation of a cytoplasmic loop to enhance nucleophilicity and modification of a nearby cysteine (C665), thereby stabilizing the loop in an activating configuration. These actions modulate two restrictions controlling ion permeation, including widening of the selectivity filter to enhance calcium permeability and opening of a canonical gate at the cytoplasmic end of the pore. We propose a model to explain functional coupling between electrophile action and these control points. We also characterize a calcium-binding pocket that is highly conserved across TRP channel subtypes and accounts for all aspects of calcium-dependent TRPA1 regulation, including potentiation, desensitization and activation by metabotropic receptors. These findings provide a structural framework for understanding how a broad-spectrum irritant receptor is controlled by endogenous and exogenous agents that elicit or exacerbate pain and itch. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21131.map.gz emd_21131.map.gz | 31.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21131-v30.xml emd-21131-v30.xml emd-21131.xml emd-21131.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
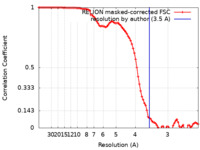
| FSC (resolution estimation) |  emd_21131_fsc.xml emd_21131_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_21131.png emd_21131.png | 90.2 KB | ||
| Masks |  emd_21131_msk_1.map emd_21131_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_21131_half_map_1.map.gz emd_21131_half_map_1.map.gz emd_21131_half_map_2.map.gz emd_21131_half_map_2.map.gz | 58.9 MB 58.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21131 http://ftp.pdbj.org/pub/emdb/structures/EMD-21131 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21131 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21131 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21131.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21131.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2156 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21131_msk_1.map emd_21131_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_21131_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_21131_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TRPA1 treated with A-967079 (but ligand not bound), solubilized i...
| Entire | Name: TRPA1 treated with A-967079 (but ligand not bound), solubilized in PMAL-C8 |
|---|---|
| Components |
|
-Supramolecule #1: TRPA1 treated with A-967079 (but ligand not bound), solubilized i...
| Supramolecule | Name: TRPA1 treated with A-967079 (but ligand not bound), solubilized in PMAL-C8 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant strain: HEK293F Homo sapiens (human) / Recombinant strain: HEK293F |
-Macromolecule #1: TRPA1
| Macromolecule | Name: TRPA1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKRSLRKMWR PGEKKEPQGV VYEDVPDDTE DFKESLKVVF EGSAYGLQNF NKQKKLKRCD DMDTFFLHYA AAEGQIELME KITRDSSLEV LHEMDDYGNT PLHCAVEKNQ IESVKFLLSR GANPNLRNFN MMAPLHIAVQ GMNNEVMKVL LEHRTIDVNL EGENGNTAVI ...String: MKRSLRKMWR PGEKKEPQGV VYEDVPDDTE DFKESLKVVF EGSAYGLQNF NKQKKLKRCD DMDTFFLHYA AAEGQIELME KITRDSSLEV LHEMDDYGNT PLHCAVEKNQ IESVKFLLSR GANPNLRNFN MMAPLHIAVQ GMNNEVMKVL LEHRTIDVNL EGENGNTAVI IACTTNNSEA LQILLKKGAK PCKSNKWGCF PIHQAAFSGS KECMEIILRF GEEHGYSRQL HINFMNNGKA TPLHLAVQNG DLEMIKMCLD NGAQIDPVEK GRCTAIHFAA TQGATEIVKL MISSYSGSVD IVNTTDGCHE TMLHRASLFD HHELADYLIS VGADINKIDS EGRSPLILAT ASASWNIVNL LLSKGAQVDI KDNFGRNFLH LTVQQPYGLK NLRPEFMQMQ QIKELVMDED NDGCTPLHYA CRQGGPGSVN NLLGFNVSIH SKSKDKKSPL HFAASYGRIN TCQRLLQDIS DTRLLNEGDL HGMTPLHLAA KNGHDKVVQL LLKKGALFLS DHNGWTALHH ASMGGYTQTM KVILDTNLKC TDRLDEDGNT ALHFAAREGH AKAVALLLSH NADIVLNKQQ ASFLHLALHN KRKEVVLTII RSKRWDECLK IFSHNSPGNK CPITEMIEYL PECMKVLLDF CMLHSTEDKS CRDYYIEYNF KYLQCPLEFT KKTPTQDVIY EPLTALNAMV QNNRIELLNH PVCKEYLLMK WLAYGFRAHM MNLGSYCLGL IPMTILVVNI KPGMAFNSTG IINETSDHSE ILDTTNSYLI KTCMILVFLS SIFGYCKEAG QIFQQKRNYF MDISNVLEWI IYTTGIIFVL PLFVEIPAHL QWQCGAIAVY FYWMNFLLYL QRFENCGIFI VMLEVILKTL LRSTVVFIFL LLAFGLSFYI LLNLQDPFSS PLLSIIQTFS MMLGDINYRE SFLEPYLRNE LAHPVLSFAQ LVSFTIFVPI VLMNLLIGLA VGDIADVQKH ASLKRIAMQV ELHTSLEKKL PLWFLRKVDQ KSTIVYPNKP RSGGMLFHIF CFLFCTGEIR QEIPNADKSL EMEILKQKYR LKDLTFLLEK QHELIKLIIQ KMEIISETED DDSHCSFQDR FKKEQMEQRN SRWNTVLRAV KAKTHHLEP |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)