+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21062 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
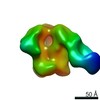
| Title | RM19A1 Fab in complex with BG505 SOSIPv5.2 | ||||||||||||||||||
 Map data Map data | RM19A1 Fab in complex with BG505 SOSIPv5.2 | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / negative staining / Resolution: 22.0 Å | ||||||||||||||||||
 Authors Authors | Cottrell CA / Ozorowski G / Ward AB | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2020 Journal: PLoS Pathog / Year: 2020Title: Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates. Authors: Christopher A Cottrell / Jelle van Schooten / Charles A Bowman / Meng Yuan / David Oyen / Mia Shin / Robert Morpurgo / Patricia van der Woude / Mariëlle van Breemen / Jonathan L Torres / ...Authors: Christopher A Cottrell / Jelle van Schooten / Charles A Bowman / Meng Yuan / David Oyen / Mia Shin / Robert Morpurgo / Patricia van der Woude / Mariëlle van Breemen / Jonathan L Torres / Raj Patel / Justin Gross / Leigh M Sewall / Jeffrey Copps / Gabriel Ozorowski / Bartek Nogal / Devin Sok / Eva G Rakasz / Celia Labranche / Vladimir Vigdorovich / Scott Christley / Diane G Carnathan / D Noah Sather / David Montefiori / Guido Silvestri / Dennis R Burton / John P Moore / Ian A Wilson / Rogier W Sanders / Andrew B Ward / Marit J van Gils /   Abstract: The induction of broad and potent immunity by vaccines is the key focus of research efforts aimed at protecting against HIV-1 infection. Soluble native-like HIV-1 envelope glycoproteins have shown ...The induction of broad and potent immunity by vaccines is the key focus of research efforts aimed at protecting against HIV-1 infection. Soluble native-like HIV-1 envelope glycoproteins have shown promise as vaccine candidates as they can induce potent autologous neutralizing responses in rabbits and non-human primates. In this study, monoclonal antibodies were isolated and characterized from rhesus macaques immunized with the BG505 SOSIP.664 trimer to better understand vaccine-induced antibody responses. Our studies reveal a diverse landscape of antibodies recognizing immunodominant strain-specific epitopes and non-neutralizing neo-epitopes. Additionally, we isolated a subset of mAbs against an epitope cluster at the gp120-gp41 interface that recognize the highly conserved fusion peptide and the glycan at position 88 and have characteristics akin to several human-derived broadly neutralizing antibodies. #1:  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: Mapping the immunogenic landscape of native HIV-1 envelope trimers in non-human primates Authors: Cottrell CA / van Schooten J / Bowman CA / Yuan M / Oyen D / Shin M / Morpurgo R / van der Woude P / van Breemen M / Torres JL / Patel R / Gross J / Sewall LM / Copps J / Ozorowski G / Sok D ...Authors: Cottrell CA / van Schooten J / Bowman CA / Yuan M / Oyen D / Shin M / Morpurgo R / van der Woude P / van Breemen M / Torres JL / Patel R / Gross J / Sewall LM / Copps J / Ozorowski G / Sok D / Rakasz EG / Labranche C / Vigdorovich V / Christley S / Sather DN / Montefiori D / Moore JP / Burton DR / Wilson IA / Sanders RW / Ward AB / van Gils MJ | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21062.map.gz emd_21062.map.gz | 20.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21062-v30.xml emd-21062-v30.xml emd-21062.xml emd-21062.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21062.png emd_21062.png | 30.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21062 http://ftp.pdbj.org/pub/emdb/structures/EMD-21062 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21062 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21062 | HTTPS FTP |
-Validation report
| Summary document |  emd_21062_validation.pdf.gz emd_21062_validation.pdf.gz | 77.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21062_full_validation.pdf.gz emd_21062_full_validation.pdf.gz | 76.8 KB | Display | |
| Data in XML |  emd_21062_validation.xml.gz emd_21062_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21062 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21062 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21062 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21062 | HTTPS FTP |
-Related structure data
| Related structure data |  6vlrC  6vn0C  6vo1C  6vorC  6vosC  6vsrC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21062.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21062.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RM19A1 Fab in complex with BG505 SOSIPv5.2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
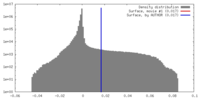
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : RM19N Fab in complex with BG505 SOSIPv5.2
| Entire | Name: RM19N Fab in complex with BG505 SOSIPv5.2 |
|---|---|
| Components |
|
-Supramolecule #1: RM19N Fab in complex with BG505 SOSIPv5.2
| Supramolecule | Name: RM19N Fab in complex with BG505 SOSIPv5.2 / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293F Homo sapiens (human) / Recombinant cell: HEK293F |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
| Grid | Support film - Material: CELLULOSE ACETATE / Details: unspecified |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 27312 |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 22.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.0) / Number images used: 6311 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera










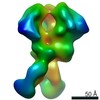



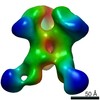
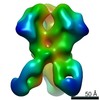

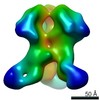


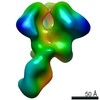

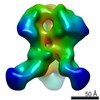


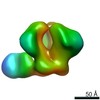















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)