[English] 日本語
 Yorodumi
Yorodumi- EMDB-21019: Cryo-EM Structure of the Hyperpolarization-Activated Potassium Ch... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21019 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the Hyperpolarization-Activated Potassium Channel KAT1: Octamer | |||||||||
 Map data Map data | Structure of the Hyperpolarization-Activated Potassium Channel KAT1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein / voltage-gated ion channel / potassium channel / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationinward rectifier potassium channel activity / monoatomic ion channel complex / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
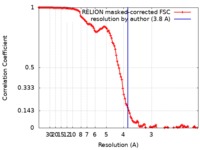
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Clark MD / Contreras GF | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Electromechanical coupling in the hyperpolarization-activated K channel KAT1. Authors: Michael David Clark / Gustavo F Contreras / Rong Shen / Eduardo Perozo /  Abstract: Voltage-gated potassium (K) channels coordinate electrical signalling and control cell volume by gating in response to membrane depolarization or hyperpolarization. However, although voltage-sensing ...Voltage-gated potassium (K) channels coordinate electrical signalling and control cell volume by gating in response to membrane depolarization or hyperpolarization. However, although voltage-sensing domains transduce transmembrane electric field changes by a common mechanism involving the outward or inward translocation of gating charges, the general determinants of channel gating polarity remain poorly understood. Here we suggest a molecular mechanism for electromechanical coupling and gating polarity in non-domain-swapped K channels on the basis of the cryo-electron microscopy structure of KAT1, the hyperpolarization-activated K channel from Arabidopsis thaliana. KAT1 displays a depolarized voltage sensor, which interacts with a closed pore domain directly via two interfaces and indirectly via an intercalated phospholipid. Functional evaluation of KAT1 structure-guided mutants at the sensor-pore interfaces suggests a mechanism in which direct interaction between the sensor and the C-linker hairpin in the adjacent pore subunit is the primary determinant of gating polarity. We suggest that an inward motion of the S4 sensor helix of approximately 5-7 Å can underlie a direct-coupling mechanism, driving a conformational reorientation of the C-linker and ultimately opening the activation gate formed by the S6 intracellular bundle. This direct-coupling mechanism contrasts with allosteric mechanisms proposed for hyperpolarization-activated cyclic nucleotide-gated channels, and may represent an unexpected link between depolarization- and hyperpolarization-activated channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21019.map.gz emd_21019.map.gz | 15.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21019-v30.xml emd-21019-v30.xml emd-21019.xml emd-21019.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21019_fsc.xml emd_21019_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_21019.png emd_21019.png | 36.9 KB | ||
| Filedesc metadata |  emd-21019.cif.gz emd-21019.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21019 http://ftp.pdbj.org/pub/emdb/structures/EMD-21019 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21019 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21019 | HTTPS FTP |
-Related structure data
| Related structure data |  6v1yMC  6v1xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-11054 (Title: Cryo-EM Structure of the Hyperpolarization-Activated Potassium Channel KAT1 EMPIAR-11054 (Title: Cryo-EM Structure of the Hyperpolarization-Activated Potassium Channel KAT1Data size: 3.1 TB Data #1: unaligned non-gain-ref movies [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21019.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21019.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the Hyperpolarization-Activated Potassium Channel KAT1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.064 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
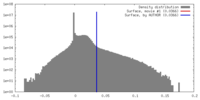
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Hyperpolarization-Activated Potassium Channel KAT1
| Entire | Name: Hyperpolarization-Activated Potassium Channel KAT1 |
|---|---|
| Components |
|
-Supramolecule #1: Hyperpolarization-Activated Potassium Channel KAT1
| Supramolecule | Name: Hyperpolarization-Activated Potassium Channel KAT1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Potassium channel KAT1
| Macromolecule | Name: Potassium channel KAT1 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 59.639594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSISWTRNFF ERFCVEEYNI DTIKQSSFLS ADLLPSLGAR INQSTKLRKH IISPFNPRYR AWEMWLVLLV IYSAWICPFQ FAFITYKKD AIFIIDNIVN GFFAIDIILT FFVAYLDSHS YLLVDSPKKI AIRYLSTWFA FDVCSTAPFQ PLSLLFNYNG S ELGFRILS ...String: MSISWTRNFF ERFCVEEYNI DTIKQSSFLS ADLLPSLGAR INQSTKLRKH IISPFNPRYR AWEMWLVLLV IYSAWICPFQ FAFITYKKD AIFIIDNIVN GFFAIDIILT FFVAYLDSHS YLLVDSPKKI AIRYLSTWFA FDVCSTAPFQ PLSLLFNYNG S ELGFRILS MLRLWRLRRV SSLFARLEKD IRFNYFWIRC TKLISVTLFA IHCAGCFNYL IADRYPNPRK TWIGAVYPNF KE ASLWNRY VTALYWSITT LTTTGYGDFH AENPREMLFD IFFMMFNLGL TAYLIGNMTN LVVHWTSRTR TFRDSVRAAS EFA SRNQLP HDIQDQMLSH ICLKFKTEGL KQQETLNNLP KAIRSSIANY LFFPIVHNIY LFQGVSRNFL FQLVSDIDAE YFPP KEDII LQNEAPTDLY ILVSGAVDFT VYVDGHDQFQ GKAVIGETFG EVGVLYYRPQ PFTVRTTELS QILRISRTSL MSAMH AHAD DGRVIMNNLF MKLRGQQSSR GSLEVLFQ UniProtKB: Potassium channel KAT1 |
-Macromolecule #2: (2S)-1-(nonanoyloxy)-3-(phosphonooxy)propan-2-yl tetradecanoate
| Macromolecule | Name: (2S)-1-(nonanoyloxy)-3-(phosphonooxy)propan-2-yl tetradecanoate type: ligand / ID: 2 / Number of copies: 8 / Formula: QNP |
|---|---|
| Molecular weight | Theoretical: 522.652 Da |
| Chemical component information |  ChemComp-QNP: |
-Macromolecule #3: (3beta,5beta,14beta,17alpha)-cholestan-3-ol
| Macromolecule | Name: (3beta,5beta,14beta,17alpha)-cholestan-3-ol / type: ligand / ID: 3 / Number of copies: 8 / Formula: QNJ |
|---|---|
| Molecular weight | Theoretical: 388.669 Da |
| Chemical component information | 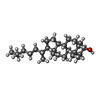 ChemComp-QNJ: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 1502 / Average exposure time: 12.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6v1y: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)