+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21015 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Two-pore channel 3 | |||||||||
 Map data Map data | Unsharpened map from non-uniform refinement in cryoSPARC v2. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Voltag-gated ion channel / two-pore channel / TPC3 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsodium ion transmembrane transporter activity / voltage-gated sodium channel activity / action potential / sodium ion transmembrane transport / protein homodimerization activity / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.11 Å | |||||||||
 Authors Authors | Dickinson MS / Stroud RM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Resting state structure of the hyperdepolarization activated two-pore channel 3. Authors: Miles Sasha Dickinson / Alexander Myasnikov / Jacob Eriksen / Nicole Poweleit / Robert M Stroud /  Abstract: Voltage-gated ion channels endow membranes with excitability and the means to propagate action potentials that form the basis of all neuronal signaling. We determined the structure of a voltage-gated ...Voltage-gated ion channels endow membranes with excitability and the means to propagate action potentials that form the basis of all neuronal signaling. We determined the structure of a voltage-gated sodium channel, two-pore channel 3 (TPC3), which generates ultralong action potentials. TPC3 is distinguished by activation only at extreme membrane depolarization (V ∼ +75 mV), in contrast to other TPCs and Na channels that activate between -20 and 0 mV. We present electrophysiological evidence that TPC3 voltage activation depends only on voltage sensing domain 2 (VSD2) and that each of the three gating arginines in VSD2 reduces the activation threshold. The structure presents a chemical basis for sodium selectivity, and a constricted gate suggests a closed pore consistent with extreme voltage dependence. The structure, confirmed by our electrophysiology, illustrates the configuration of a bona fide resting state voltage sensor, observed without the need for any inhibitory ligand, and independent of any chemical or mutagenic alteration. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21015.map.gz emd_21015.map.gz | 41.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21015-v30.xml emd-21015-v30.xml emd-21015.xml emd-21015.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21015.png emd_21015.png | 200.7 KB | ||
| Filedesc metadata |  emd-21015.cif.gz emd-21015.cif.gz | 5.8 KB | ||
| Others |  emd_21015_additional.map.gz emd_21015_additional.map.gz | 79 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21015 http://ftp.pdbj.org/pub/emdb/structures/EMD-21015 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21015 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21015 | HTTPS FTP |
-Related structure data
| Related structure data |  6v1qMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21015.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21015.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map from non-uniform refinement in cryoSPARC v2. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
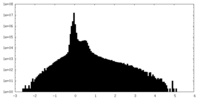
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.256 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
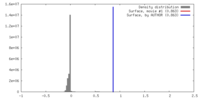
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Locally sharpened map from non-uniform refinement in cryoSPARC...
| File | emd_21015_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally sharpened map from non-uniform refinement in cryoSPARC v2, used for atomic modeling. | ||||||||||||
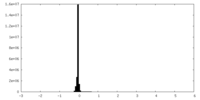
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dimer of two-pore channel 3 subunits
| Entire | Name: Dimer of two-pore channel 3 subunits |
|---|---|
| Components |
|
-Supramolecule #1: Dimer of two-pore channel 3 subunits
| Supramolecule | Name: Dimer of two-pore channel 3 subunits / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Two pore channel 3
| Macromolecule | Name: Two pore channel 3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 88.774164 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSEGKTEKTS HTLTKDEGFT NGGNHVPSNV TDQMTEKFDL ATVYVSDAKY NRNIFFDTSP QAVKLYLLYN HWFMQTLVYV FIIINLALA LFEDPAVVPL PIWATSTIET ICLSAFTVRI IHYAKVIPKD KFWKDPKNIC IIIIVTLSFI DMVIYGALKA T GHYGIRWS ...String: MSEGKTEKTS HTLTKDEGFT NGGNHVPSNV TDQMTEKFDL ATVYVSDAKY NRNIFFDTSP QAVKLYLLYN HWFMQTLVYV FIIINLALA LFEDPAVVPL PIWATSTIET ICLSAFTVRI IHYAKVIPKD KFWKDPKNIC IIIIVTLSFI DMVIYGALKA T GHYGIRWS RVLRPLLLVN VTEGRQLRRA FRSIRNALPQ ISYVFFLFMF SVLVFSLMAL KLFGKRGLLT INGSPYFTDY MD IVFDLYV LVTTANSPDV MMPAYNSSVY FTIFFILYIV INTYTFMSFF LAVVYNNYKK YLKEEVRQLV KAKRIKMCRA FSL LQENRG EGGEPVVTQA NWNHLVKLVK PKISTAHREL LWSVLDDQNK GHIGKFAFVQ LADLLSIQVI TVKSQAHPIQ ICFP SLYNS LPSRFIRQMV HHRVFVYAYD LIILVNAVFI GLDEENPVVS NAEWGFLALY MLEILLKLYA TEPRAFFARH QFWNW FDTI IVVSALFGTI INSALKHSGG YTSRQVLDIV FILRVLRLIR VVDSIKRFRA IINTLIKIGP TILTFGQLIL VVYYIF AMV GMELFKGKIQ FFEPNSTSPD REYCGNPLLK STSFAKLNYC KNNFNDVISS FILLLELTVV NQWHVLTSGF TAVTHVS AR LFFVIFHIVV VIIIINIFVA FILEAFLVEY TVDKSELQTS LEKKIEELEL NVQQDGVDTG LVDAMETNDS DLGSSEDG K RKPSLMFKIA SRRSRTVDGL LQRMFETDLR PEDFNEEELD NTNFSNPVFD SV UniProtKB: Two pore channel 3 |
-Macromolecule #2: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 2 / Number of copies: 1 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 92.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.11 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 2) / Number images used: 213328 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)