[English] 日本語
 Yorodumi
Yorodumi- EMDB-21006: Negative stain EM map of CODH/ACS in the hyperextended conformation -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21006 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
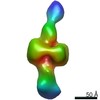
| Title | Negative stain EM map of CODH/ACS in the hyperextended conformation | |||||||||||||||
 Map data Map data | Negative stain electron microscopy map of CODH/ACS in the hyperextended state | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  Moorella thermoacetica (bacteria) Moorella thermoacetica (bacteria) | |||||||||||||||
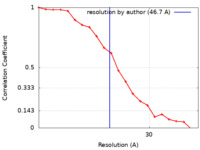
| Method | single particle reconstruction / negative staining / Resolution: 46.7 Å | |||||||||||||||
 Authors Authors | Cohen SE / Brignole EJ / Drennan CL | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2021 Journal: Structure / Year: 2021Title: Negative-Stain Electron Microscopy Reveals Dramatic Structural Rearrangements in Ni-Fe-S-Dependent Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase. Authors: Steven E Cohen / Edward J Brignole / Elizabeth C Wittenborn / Mehmet Can / Samuel Thompson / Stephen W Ragsdale / Catherine L Drennan /  Abstract: The Ni-Fe-S-containing A-cluster of acetyl-coenzyme A (CoA) synthase (ACS) assembles acetyl-CoA from carbon monoxide (CO), a methyl group (CH), and CoA. To accomplish this feat, ACS must bind CoA and ...The Ni-Fe-S-containing A-cluster of acetyl-coenzyme A (CoA) synthase (ACS) assembles acetyl-CoA from carbon monoxide (CO), a methyl group (CH), and CoA. To accomplish this feat, ACS must bind CoA and interact with two other proteins that contribute the CO and CH, respectively: CO dehydrogenase (CODH) and corrinoid Fe-S protein (CFeSP). Previous structural data show that, in the model acetogen Moorella thermoacetica, domain 1 of ACS binds to CODH such that a 70-Å-long internal channel is created that allows CO to travel from CODH to the A-cluster. The A-cluster is largely buried and is inaccessible to CFeSP for methylation. Here we use electron microscopy to capture multiple snapshots of ACS that reveal previously uncharacterized domain motion, forming extended and hyperextended structural states. In these structural states, the A-cluster is accessible for methylation by CFeSP. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21006.map.gz emd_21006.map.gz | 938.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21006-v30.xml emd-21006-v30.xml emd-21006.xml emd-21006.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21006_fsc.xml emd_21006_fsc.xml | 1.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_21006.png emd_21006.png | 29.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21006 http://ftp.pdbj.org/pub/emdb/structures/EMD-21006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21006 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21006 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21006.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21006.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain electron microscopy map of CODH/ACS in the hyperextended state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.16 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : carbon monoxide dheydrogenase/acetyl-CoA synthase tetramer
| Entire | Name: carbon monoxide dheydrogenase/acetyl-CoA synthase tetramer |
|---|---|
| Components |
|
-Supramolecule #1: carbon monoxide dheydrogenase/acetyl-CoA synthase tetramer
| Supramolecule | Name: carbon monoxide dheydrogenase/acetyl-CoA synthase tetramer type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Moorella thermoacetica (bacteria) Moorella thermoacetica (bacteria) |
-Macromolecule #1: Carbon monoxide dehydrogenase
| Macromolecule | Name: Carbon monoxide dehydrogenase / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: anaerobic carbon monoxide dehydrogenase |
|---|---|
| Source (natural) | Organism:  Moorella thermoacetica (bacteria) Moorella thermoacetica (bacteria) |
| Sequence | String: MPRFRDLSHN CRPSEAPRVM EPKNRDRTVD PAVLEMLVKS KDDKVITAFD RFVAQQPQCK IGYEGICCRF CMAGPCRIKA TDGPGSRGIC GASAWTIVAR NVGLMILTGA AAHCEHGNHI AHALVEMAEG KAPDYSVKDE AKLKEVCRRV GIEVEGKSVL ELAQEVGEKA ...String: MPRFRDLSHN CRPSEAPRVM EPKNRDRTVD PAVLEMLVKS KDDKVITAFD RFVAQQPQCK IGYEGICCRF CMAGPCRIKA TDGPGSRGIC GASAWTIVAR NVGLMILTGA AAHCEHGNHI AHALVEMAEG KAPDYSVKDE AKLKEVCRRV GIEVEGKSVL ELAQEVGEKA LEDFRRLKGE GEATWLMTTI NEGRKEKFRT HNVVPFGIHA SISELVNQAH MGMDNDPVNL VFSAIRVALA DYTGEHIATD FSDILFGTPQ PVVSEANMGV LDPDQVNFVL HGHNPLLSEI IVQAAREMEG EAKAAGAKGI NLVGICCTGN EVLMRQGIPL VTSFASQELA ICTGAIDAMC VDVQCIMPSI SAVAECYHTR IITTADNAKI PGAYHIDYQT ATAIESAKTA IRMAIEAFKE RKESNRPVYI PQIKNRVVAG WSLEALTKLL ATQNAQNPIR VLNQAILDGE LAGVALICGC NNLKGFQDNS HLTVMKELLK NNVFVVATGC SAQAAGKLGL LDPANVETYC GDGLKGFLKR LGEGANIEIG LPPVFHMGSC VDNSRAVDLL MAMANDLGVD TPKVPFVASA PEAMSGKAAA IGTWWVSLGV PTHVGTMPPV EGSDLIYSIL TQIASDVYGG YFIFEMDPQV AARKILDALE YRTWKLGVHK EVAERYETKL CQGY |
-Macromolecule #2: Acetyl-CoA synthase
| Macromolecule | Name: Acetyl-CoA synthase / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO / EC number: CO-methylating acetyl-CoA synthase |
|---|---|
| Source (natural) | Organism:  Moorella thermoacetica (bacteria) Moorella thermoacetica (bacteria) |
| Sequence | String: MTDFDKIFEG AIPEGKEPVA LFREVYHGAI TATSYAEILL NQAIRTYGPD HPVGYPDTAY YLPVIRCFSG EEVKKLGDLP PILNRKRAQV SPVLNFENAR LAGEATWYAA EIIEALRYLK YKPDEPLLPP PWTGFIGDPV VRRFGIKMVD WTIPGEAIIL GRAKDSKALA ...String: MTDFDKIFEG AIPEGKEPVA LFREVYHGAI TATSYAEILL NQAIRTYGPD HPVGYPDTAY YLPVIRCFSG EEVKKLGDLP PILNRKRAQV SPVLNFENAR LAGEATWYAA EIIEALRYLK YKPDEPLLPP PWTGFIGDPV VRRFGIKMVD WTIPGEAIIL GRAKDSKALA KIVKELMGMG FMLFICDEAV EQLLEENVKL GIDYIAYPLG NFTQIVHAAN YALRAGMMFG GVTPGAREEQ RDYQRRRIRA FVLYLGEHDM VKTAAAFGAI FTGFPVITDQ PLPEDKQIPD WFFSVEDYDK IVQIAMETRG IKLTKIKLDL PINFGPAFEG ESIRKGDMYV EMGGNRTPAF ELVRTVSESE ITDGKIEVIG PDIDQIPEGS KLPLGILVDI YGRKMQADFE GVLERRIHDF INYGEGLWHT GQRNINWLRV SKDAVAKGFR FKNYGEILVA KMKEEFPAIV DRVQVTIFTD EAKVKEYMEV AREKYKERDD RMRGLTDETV DTFYSCVLCQ SFAPNHVCIV TPERVGLCGA VSWLDAKASY EINHAGPNQP IPKEGEIDPI KGIWKSVNDY LYTASNRNLE QVCLYTLMEN PMTSCGCFEA IMAILPECNG IMITTRDHAG MTPSGMTFST LAGMIGGGTQ TPGFMGIGRT YIVSKKFISA DGGIARIVWM PKSLKDFLHD EFVRRSVEEG LGEDFIDKIA DETIGTTVDE ILPYLEEKGH PALTMDPIM |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | .017 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| |||||||||
| Staining | Type: NEGATIVE / Material: uranyl acetate Details: Samples were prepared by blotting protein from EM grid using filter paper, followed by multiple rounds of staining and blotting. | |||||||||
| Grid | Support film - Material: CARBON / Support film - topology: CONTINUOUS / Details: unspecified | |||||||||
| Details | The sample was predominantly monodisperse, as seen by negative-stain TEM. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number grids imaged: 1 / Average exposure time: 1.0 sec. / Average electron dose: 23.0 e/Å2 Details: Images were collected as tilt pairs at 0 and 55 degrees. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal magnification: 62000 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)