+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20843 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Alpha-E-catenin ABD-F-actin complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | alpha-catenin / catenin / actin / mechanobiology / mechanosensing / cytoskeleton / cell adhesion | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of integrin-mediated signaling pathway / CDH11 homotypic and heterotypic interactions / Regulation of CDH19 Expression and Function / Regulation of CDH11 function / gamma-catenin binding / epithelial cell-cell adhesion / zonula adherens / gap junction assembly / Striated Muscle Contraction / cellular response to indole-3-methanol ...negative regulation of integrin-mediated signaling pathway / CDH11 homotypic and heterotypic interactions / Regulation of CDH19 Expression and Function / Regulation of CDH11 function / gamma-catenin binding / epithelial cell-cell adhesion / zonula adherens / gap junction assembly / Striated Muscle Contraction / cellular response to indole-3-methanol / flotillin complex / vinculin binding / catenin complex / apical junction assembly / negative regulation of cell motility / positive regulation of extrinsic apoptotic signaling pathway in absence of ligand / positive regulation of smoothened signaling pathway / Adherens junctions interactions / negative regulation of protein localization to nucleus / axon regeneration / negative regulation of neuroblast proliferation / smoothened signaling pathway / Myogenesis / odontogenesis of dentin-containing tooth / establishment or maintenance of cell polarity / striated muscle thin filament / skeletal muscle thin filament assembly / intercalated disc / neuroblast proliferation / negative regulation of extrinsic apoptotic signaling pathway in absence of ligand / RHO GTPases activate IQGAPs / ovarian follicle development / skeletal muscle fiber development / stress fiber / extrinsic apoptotic signaling pathway in absence of ligand / acrosomal vesicle / VEGFR2 mediated vascular permeability / integrin-mediated signaling pathway / actin filament / adherens junction / cell-cell adhesion / beta-catenin binding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / response to estrogen / male gonad development / actin filament binding / cell-cell junction / cell junction / intracellular protein localization / cell migration / lamellipodium / actin cytoskeleton / cell adhesion / hydrolase activity / cadherin binding / focal adhesion / intracellular membrane-bounded organelle / structural molecule activity / Golgi apparatus / RNA binding / ATP binding / identical protein binding / nucleus / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
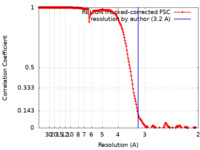
| Method | helical reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Mei L / Alushin GM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Molecular mechanism for direct actin force-sensing by α-catenin. Authors: Lin Mei / Santiago Espinosa de Los Reyes / Matthew J Reynolds / Rachel Leicher / Shixin Liu / Gregory M Alushin /  Abstract: The actin cytoskeleton mediates mechanical coupling between cells and their tissue microenvironments. The architecture and composition of actin networks are modulated by force; however, it is unclear ...The actin cytoskeleton mediates mechanical coupling between cells and their tissue microenvironments. The architecture and composition of actin networks are modulated by force; however, it is unclear how interactions between actin filaments (F-actin) and associated proteins are mechanically regulated. Here we employ both optical trapping and biochemical reconstitution with myosin motor proteins to show single piconewton forces applied solely to F-actin enhance binding by the human version of the essential cell-cell adhesion protein αE-catenin but not its homolog vinculin. Cryo-electron microscopy structures of both proteins bound to F-actin reveal unique rearrangements that facilitate their flexible C-termini refolding to engage distinct interfaces. Truncating α-catenin's C-terminus eliminates force-activated F-actin binding, and addition of this motif to vinculin confers force-activated binding, demonstrating that α-catenin's C-terminus is a modular detector of F-actin tension. Our studies establish that piconewton force on F-actin can enhance partner binding, which we propose mechanically regulates cellular adhesion through α-catenin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20843.map.gz emd_20843.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20843-v30.xml emd-20843-v30.xml emd-20843.xml emd-20843.xml | 26.9 KB 26.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20843_fsc.xml emd_20843_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_20843.png emd_20843.png | 123.4 KB | ||
| Masks |  emd_20843_msk_1.map emd_20843_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20843.cif.gz emd-20843.cif.gz | 7.8 KB | ||
| Others |  emd_20843_additional_1.map.gz emd_20843_additional_1.map.gz emd_20843_additional_2.map.gz emd_20843_additional_2.map.gz emd_20843_half_map_1.map.gz emd_20843_half_map_1.map.gz emd_20843_half_map_2.map.gz emd_20843_half_map_2.map.gz | 282.9 MB 408.1 MB 410.7 MB 410.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20843 http://ftp.pdbj.org/pub/emdb/structures/EMD-20843 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20843 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20843 | HTTPS FTP |
-Validation report
| Summary document |  emd_20843_validation.pdf.gz emd_20843_validation.pdf.gz | 777.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20843_full_validation.pdf.gz emd_20843_full_validation.pdf.gz | 777.8 KB | Display | |
| Data in XML |  emd_20843_validation.xml.gz emd_20843_validation.xml.gz | 23.3 KB | Display | |
| Data in CIF |  emd_20843_validation.cif.gz emd_20843_validation.cif.gz | 29.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20843 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20843 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20843 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20843 | HTTPS FTP |
-Related structure data
| Related structure data |  6upvMC  6upwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10654 (Title: Alpha-E-catenin ABD-F-actin complex / Data size: 2.4 TB EMPIAR-10654 (Title: Alpha-E-catenin ABD-F-actin complex / Data size: 2.4 TBData #1: unaligned multi-frame micrographs of alpha-catenin ABD-actin complex [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20843.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20843.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20843_msk_1.map emd_20843_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
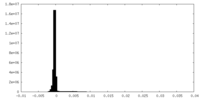
| Density Histograms |
-Additional map: B-factor sharpened, local resolution-filtered map
| File | emd_20843_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor sharpened, local resolution-filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
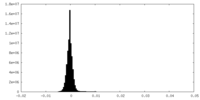
| Density Histograms |
-Additional map: unfiltered, unsharpened map
| File | emd_20843_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered, unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_20843_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
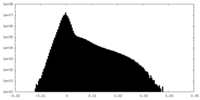
| Density Histograms |
-Half map: #2
| File | emd_20843_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
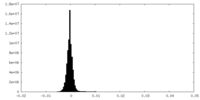
| Density Histograms |
- Sample components
Sample components
-Entire : alpha-E-catenin ABD-F-actin complex
| Entire | Name: alpha-E-catenin ABD-F-actin complex |
|---|---|
| Components |
|
-Supramolecule #1: alpha-E-catenin ABD-F-actin complex
| Supramolecule | Name: alpha-E-catenin ABD-F-actin complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Actin binding domain (residues 664-906) of alpha-E-catenin bound to F-actin |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.5 kDa/nm |
-Macromolecule #1: Catenin alpha-1
| Macromolecule | Name: Catenin alpha-1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 100.206352 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTAVHAGNIN FKWDPKSLEI RTLAVERLLE PLVTQVTTLV NTNSKGPSNK KRGRSKKAHV LAASVEQATE NFLEKGDKIA KESQFLKEE LVAAVEDVRK QGDLMKAAAG EFADDPCSSV KRGNMVRAAR ALLSAVTRLL ILADMADVYK LLVQLKVVED G ILKLRNAG ...String: MTAVHAGNIN FKWDPKSLEI RTLAVERLLE PLVTQVTTLV NTNSKGPSNK KRGRSKKAHV LAASVEQATE NFLEKGDKIA KESQFLKEE LVAAVEDVRK QGDLMKAAAG EFADDPCSSV KRGNMVRAAR ALLSAVTRLL ILADMADVYK LLVQLKVVED G ILKLRNAG NEQDLGIQYK ALKPEVDKLN IMAAKRQQEL KDVGHRDQMA AARGILQKNV PILYTASQAC LQHPDVAAYK AN RDLIYKQ LQQAVTGISN AAQATASDDA SQHQGGGGGE LAYALNNFDK QIIVDPLSFS EERFRPSLEE RLESIISGAA LMA DSSCTR DDRRERIVAE CNAVRQALQD LLSEYMGNAG RKERSDALNS AIDKMTKKTR DLRRQLRKAV MDHVSDSFLE TNVP LLVLI EAAKNGNEKE VKEYAQVFRE HANKLIEVAN LACSISNNEE GVKLVRMSAS QLEALCPQVI NAALALAAKP QSKLA QENM DLFKEQWEKQ VRVLTDAVDD ITSIDDFLAV SENHILEDVN KCVIALQEKD VDGLDRTAGA IRGRAARVIH VVTSEM DNY EPGVYTEKVL EATKLLSNTV MPRFTEQVEA AVEALSSDPA QPMDENEFID ASRLVYDGIR DIRKAVLMIR TPEELDD SD FETEDFDVRS RTSVQTEDDQ LIAGQSARAI MAQLPQEQKA KIAEQVASFQ EEKSKLDAEV SKWDDSGNDI IVLAKQMC M IMMEMTDFTR GKGPLKNTSD VISAAKKIAE AGSRMDKLGR TIADHCPDSA CKQDLLAYLQ RIALYCHQLN ICSKVKAEV QNLGGELVVS GVDSAMSLIQ AAKNLMNAVV QTVKASYVAS TKYQKSQGMA SLNLPAVSWK MKAPEKKPLV KREKQDETQT KIKRASQKK HVNPVQALSE FKAMDSI UniProtKB: Catenin alpha-1 |
-Macromolecule #2: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.875633 KDa |
| Sequence | String: DEDETTALVC DNGSGLVKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IE(HIC)GII TNW DDMEKIWHHT FYNELRVAPE EHPTLLTEAP LNPKANREKM TQIMFETFNV PAMYVAIQAV LSLYASGRTT GIVLDSG DG VTHNVPIYEG ...String: DEDETTALVC DNGSGLVKAG FAGDDAPRAV FPSIVGRPRH QGVMVGMGQK DSYVGDEAQS KRGILTLKYP IE(HIC)GII TNW DDMEKIWHHT FYNELRVAPE EHPTLLTEAP LNPKANREKM TQIMFETFNV PAMYVAIQAV LSLYASGRTT GIVLDSG DG VTHNVPIYEG YALPHAIMRL DLAGRDLTDY LMKILTERGY SFVTTAEREI VRDIKEKLCY VALDFENEMA TAASSSSL E KSYELPDGQV ITIGNERFRC PETLFQPSFI GMESAGIHET TYNSIMKCDI DIRKDLYANN VMSGGTTMYP GIADRMQKE ITALAPSTMK IKIIAPPERK YSVWIGGSIL ASLSTFQQMW ITKQEYDEAG PSIVHRKCF UniProtKB: Actin, alpha skeletal muscle |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 5 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Average exposure time: 10.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)