[English] 日本語
 Yorodumi
Yorodumi- EMDB-20781: Structural basis for control of antibiotic production by bacteria... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20781 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural basis for control of antibiotic production by bacterial hormones | |||||||||
 Map data Map data | Map file - Postprocess masked .mrc UCSF Chimera 1.11, visualization -Volume viewer step 1 -Contour level 0.0181 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) | |||||||||
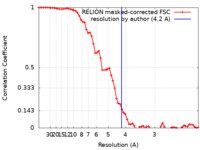
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Bhukya H | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Molecular basis for control of antibiotic production by a bacterial hormone. Authors: Shanshan Zhou / Hussain Bhukya / Nicolas Malet / Peter J Harrison / Dean Rea / Matthew J Belousoff / Hariprasad Venugopal / Paulina K Sydor / Kathryn M Styles / Lijiang Song / Max J Cryle / ...Authors: Shanshan Zhou / Hussain Bhukya / Nicolas Malet / Peter J Harrison / Dean Rea / Matthew J Belousoff / Hariprasad Venugopal / Paulina K Sydor / Kathryn M Styles / Lijiang Song / Max J Cryle / Lona M Alkhalaf / Vilmos Fülöp / Gregory L Challis / Christophe Corre /   Abstract: Actinobacteria produce numerous antibiotics and other specialized metabolites that have important applications in medicine and agriculture. Diffusible hormones frequently control the production of ...Actinobacteria produce numerous antibiotics and other specialized metabolites that have important applications in medicine and agriculture. Diffusible hormones frequently control the production of such metabolites by binding TetR family transcriptional repressors (TFTRs), but the molecular basis for this remains unclear. The production of methylenomycin antibiotics in Streptomyces coelicolor A3(2) is initiated by the binding of 2-alkyl-4-hydroxymethylfuran-3-carboxylic acid (AHFCA) hormones to the TFTR MmfR. Here we report the X-ray crystal structure of an MmfR-AHFCA complex, establishing the structural basis for hormone recognition. We also elucidate the mechanism for DNA release upon hormone binding through the single-particle cryo-electron microscopy structure of an MmfR-operator complex. DNA binding and release assays with MmfR mutants and synthetic AHFCA analogues define the role of individual amino acid residues and hormone functional groups in ligand recognition and DNA release. These findings will facilitate the exploitation of actinobacterial hormones and their associated TFTRs in synthetic biology and in the discovery of new antibiotics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20781.map.gz emd_20781.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20781-v30.xml emd-20781-v30.xml emd-20781.xml emd-20781.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20781_fsc.xml emd_20781_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_20781.png emd_20781.png | 50 KB | ||
| Masks |  emd_20781_msk_1.map emd_20781_msk_1.map | 22.2 MB |  Mask map Mask map | |
| Others |  emd_20781_half_map_1.map.gz emd_20781_half_map_1.map.gz emd_20781_half_map_2.map.gz emd_20781_half_map_2.map.gz | 17.1 MB 17.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20781 http://ftp.pdbj.org/pub/emdb/structures/EMD-20781 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20781 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20781 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20781.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20781.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map file - Postprocess masked .mrc UCSF Chimera 1.11, visualization -Volume viewer step 1 -Contour level 0.0181 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.092 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20781_msk_1.map emd_20781_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map file - run half1 class001 unfil.mrc UCSF Chimera 1.11,...
| File | emd_20781_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map file - run_half1_class001_unfil.mrc UCSF Chimera 1.11, visualization -Volume viewer step 1 -Contour level 0.017 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map file - run half2 class001 unfil.mrc UCSF Chimera 1.11,...
| File | emd_20781_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map file - run_half2_class001_unfil.mrc UCSF Chimera 1.11, visualization -Volume viewer step 1 -Contour level 0.017 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : protein-DNA complex
| Entire | Name: protein-DNA complex |
|---|---|
| Components |
|
-Supramolecule #1: protein-DNA complex
| Supramolecule | Name: protein-DNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: protein-DNA complex. two homodimers of protein bound to a double stranded DNA |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 114 KDa |
-Macromolecule #1: protein MmfR
| Macromolecule | Name: protein MmfR / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor A3(2) (bacteria) / Strain: A3(2) Streptomyces coelicolor A3(2) (bacteria) / Strain: A3(2) |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGKP IPNPLLGLDS TENLYFQGID PFTMTSAQQP TPFAVRSNVP RGPHPQQERS IKTRAQILEA ASEIFASRGY RGASVKDVAE RVGMTKGAVY FHFPSKESLA IAVVEEHYAR WPAAMEEIRI QGFTPLETVE EMLHRAAQAF RDDPVMQAGA RLQSERAFID ...String: MHHHHHHGKP IPNPLLGLDS TENLYFQGID PFTMTSAQQP TPFAVRSNVP RGPHPQQERS IKTRAQILEA ASEIFASRGY RGASVKDVAE RVGMTKGAVY FHFPSKESLA IAVVEEHYAR WPAAMEEIRI QGFTPLETVE EMLHRAAQAF RDDPVMQAGA RLQSERAFID AELPLPYVDW THLLEVPLQD AREAGQLRAG VDPAAAARSL VAAFFGMQHV SDNLHQRADI MERWQELREL MFFALRA |
-Macromolecule #2: synthetic DNA
| Macromolecule | Name: synthetic DNA / type: dna / ID: 2 / Details: 5'-ATACCTGCGGGAAGGTATT-3' / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Streptomyces coelicolor A3(2) (bacteria) Streptomyces coelicolor A3(2) (bacteria) |
| Sequence | String: ATACCTGCGG GAAGGTATT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.08 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 20 mM tris pH 8.0 and 200 mM NaCl fresh and autoclaved | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III | |||||||||
| Details | protein-DNA complex |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-50 / Number grids imaged: 2 / Average exposure time: 10.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus min: -0.0005 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT / Overall B value: 214 |
|---|
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)