+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20753 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Rabies SAD-B19 L-P complex from cryo-EM | |||||||||
 Map data Map data | Final B-factor sharpened, clipped, full EM map into which the structure was fit | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Polymerase / Phosphoprotein / Complex / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host transcription / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication / microtubule-dependent intracellular transport of viral material towards nucleus / viral transcription / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / virion component / host cell ...symbiont-mediated suppression of host transcription / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication / microtubule-dependent intracellular transport of viral material towards nucleus / viral transcription / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / virion component / host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / RNA-directed RNA polymerase / GTPase activity / RNA-directed RNA polymerase activity / symbiont entry into host cell / host cell nucleus / ATP binding Similarity search - Function | |||||||||
| Biological species |  Rabies virus (strain SAD B19) Rabies virus (strain SAD B19) | |||||||||
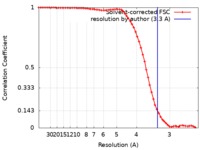
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Horwitz JA / Harrison SC | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Structure of a rabies virus polymerase complex from electron cryo-microscopy. Authors: Joshua A Horwitz / Simon Jenni / Stephen C Harrison / Sean P J Whelan /  Abstract: Nonsegmented negative-stranded (NNS) RNA viruses, among them the virus that causes rabies (RABV), include many deadly human pathogens. The large polymerase (L) proteins of NNS RNA viruses carry all ...Nonsegmented negative-stranded (NNS) RNA viruses, among them the virus that causes rabies (RABV), include many deadly human pathogens. The large polymerase (L) proteins of NNS RNA viruses carry all of the enzymatic functions required for viral messenger RNA (mRNA) transcription and replication: RNA polymerization, mRNA capping, and cap methylation. We describe here a complete structure of RABV L bound with its phosphoprotein cofactor (P), determined by electron cryo-microscopy at 3.3 Å resolution. The complex closely resembles the vesicular stomatitis virus (VSV) L-P, the one other known full-length NNS-RNA L-protein structure, with key local differences (e.g., in L-P interactions). Like the VSV L-P structure, the RABV complex analyzed here represents a preinitiation conformation. Comparison with the likely elongation state, seen in two structures of pneumovirus L-P complexes, suggests differences between priming/initiation and elongation complexes. Analysis of internal cavities within RABV L suggests distinct template and product entry and exit pathways during transcription and replication. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20753.map.gz emd_20753.map.gz | 5.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20753-v30.xml emd-20753-v30.xml emd-20753.xml emd-20753.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20753_fsc.xml emd_20753_fsc.xml | 6.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_20753.png emd_20753.png | 73.3 KB | ||
| Filedesc metadata |  emd-20753.cif.gz emd-20753.cif.gz | 6.8 KB | ||
| Others |  emd_20753_additional.map.gz emd_20753_additional.map.gz emd_20753_half_map_1.map.gz emd_20753_half_map_1.map.gz emd_20753_half_map_2.map.gz emd_20753_half_map_2.map.gz | 20.6 MB 3.7 MB 3.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20753 http://ftp.pdbj.org/pub/emdb/structures/EMD-20753 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20753 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20753 | HTTPS FTP |
-Related structure data
| Related structure data |  6uebMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20753.map.gz / Format: CCP4 / Size: 6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20753.map.gz / Format: CCP4 / Size: 6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final B-factor sharpened, clipped, full EM map into which the structure was fit | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.234 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Final unsharpened, unclipped EM map
| File | emd_20753_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final unsharpened, unclipped EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map #1
| File | emd_20753_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map #1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map #2
| File | emd_20753_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map #2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rabies lyssavirus strain SAD-B19 L-P complex
| Entire | Name: Rabies lyssavirus strain SAD-B19 L-P complex |
|---|---|
| Components |
|
-Supramolecule #1: Rabies lyssavirus strain SAD-B19 L-P complex
| Supramolecule | Name: Rabies lyssavirus strain SAD-B19 L-P complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Purified L-P(1-91) complexes from baculovirus co-expression in insect cells |
|---|---|
| Source (natural) | Organism:  Rabies virus (strain SAD B19) Rabies virus (strain SAD B19) |
-Macromolecule #1: Large structural protein
| Macromolecule | Name: Large structural protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Rabies virus (strain SAD B19) / Strain: SAD B19 Rabies virus (strain SAD B19) / Strain: SAD B19 |
| Molecular weight | Theoretical: 243.274344 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLDPGEVYDD PIDPIELEAE PRGTPIVPNI LRNSDYNLNS PLIEDPARLM LEWLKTGNRP YRMTLTDNCS RSFRVLKDYF KKVDLGSLK VGGMAAQSMI SLWLYGAHSE SNRSRRCITD LAHFYSKSSP IEKLLNLTLG NRGLRIPPEG VLSCLERVDY D NAFGRYLA ...String: MLDPGEVYDD PIDPIELEAE PRGTPIVPNI LRNSDYNLNS PLIEDPARLM LEWLKTGNRP YRMTLTDNCS RSFRVLKDYF KKVDLGSLK VGGMAAQSMI SLWLYGAHSE SNRSRRCITD LAHFYSKSSP IEKLLNLTLG NRGLRIPPEG VLSCLERVDY D NAFGRYLA NTYSSYLFFH VITLYMNALD WDEEKTILAL WKDLTSVDIG KDLVKFKDQI WGLLIVTKDF VYSQSSNCLF DR NYTLMLK DLFLSRFNSL MVLLSPPEPR YSDDLISQLC QLYIAGDQVL SMCGNSGYEV IKILEPYVVN SLVQRAEKFR PLI HSLGDF PVFIKDKVSQ LEETFGPCAR RFFRALDQFD NIHDLVFVFG CYRHWGHPYI DYRKGLSKLY DQVHLKKMID KSYQ ECLAS DLARRILRWG FDKYSKWYLD SRFLARDHPL TPYIKTQTWP PKHIVDLVGD TWHKLPITQI FEIPESMDPS EILDD KSHS FTRTRLASWL SENRGGPVPS EKVIITALSK PPVNPREFLR SIDLGGLPDE DLIIGLKPKE RELKIEGRFF ALMSWN LRL YFVITEKLLA NYILPLFDAL TMTDNLNKVF KKLIDRVTGQ GLLDYSRVTY AFHLDYEKWN NHQRLESTED VFSVLDQ VF GLKRVFSRTH EFFQKAWIYY SDRSDLIGLR EDQIYCLDAS NGPTCWNGQD GGLEGLRQKG WSLVSLLMID RESQIRNT R TKILAQGDNQ VLCPTYMLSP GLSQEGLLYE LERISRNALS IYRAVEEGAS KLGLIIKKEE TMCSYDFLIY GKTPLFRGN ILVPESKRWA RVSCVSNDQI VNLANIMSTV STNALTVAQH SQSLIKPMRD FLLMSVQAVF HYLLFSPILK GRVYKILSAE GESFLLAMS RIIYLDPSLG GISGMSLGRF HIRQFSDPVS EGLSFWREIW LSSQESWIHA LCQEAGNPDL GERTLESFTR L LEDPTTLN IRGGASPTIL LKDAIRKALY DEVDKVENSE FREAILLSKT HRDNFILFLI SVEPLFPRFL SELFSSSFLG IP ESIIGLI QNSRTIRRQF RKSLSKTLEE SFYNSEIHGI SRMTQTPQRV GGVWPCSSER ADLLREISWG RKVVGTTVPH PSE MLGLLP KSSISCTCGA TGGGNPRVSV SVLPSFDQSF FSRGPLKGYL GSSTSMSTQL FHAWEKVTNV HVVKRALSLK ESIN WFITR DSNLAQALIR NIMSLTGPDF PLEEAPVFKR TGSALHRFKS ARYSEGGYSS VCPNLLSHIS VSTDTMSDLT QDGKN YDFM FQPLMLYAQT WTSELVQRDT RLRDSTFHWH LRCNRCVRPI DDVTLETSQI FEFPDVSKRI SRMVSGAVPH FQRLPD IRL RPGDFESLSG REKSHHIGSA QGLLYSILVA IHDSGYNDGT IFPVNIYGKV SPRDYLRGLA RGVLIGSSIC FLTRMTN IN INRPLELVSG VISYILLRLD NHPSLYIMLR EPSLRGEIFS IPQKIPAAYP TTMKEGNRSI LCYLQHVLRY EREIITAS P ENDWLWIFSD FRSAKMTYLS LITYQSHLLL QRVERNLSKS MRDNLRQLSS LMRQVLGGHG EDTLESDDNI QRLLKDSLR RTRWVDQEVR HAARTMTGDY SPNKKVSRKV GCSEWVCSAQ QVAVSTSANP APVSELDIRA LSKRFQNPLI SGLRVVQWAT GAHYKLKPI LDDLNVFPSL CLVVGDGSGG ISRAVLNMFP DAKLVFNSLL EVNDLMASGT HPLPPSAIMR GGNDIVSRVI D LDSIWEKP SDLRNLATWK YFQSVQKQVN MSYDLIICDA EVTDIASINR ITLLMSDFAL SIDGPLYLVF KTYGTMLVNP NY KAIQHLS RAFPSVTGFI TQVTSSFSSE LYLRFSKRGK FFRDAEYLTS STLREMSLVL FNCSSPKSEM QRARSLNYQD LVR GFPEEI ISNPYNEMII TLIDSDVESF LVHKMVDDLE LQRGTLSKVA IIIAIMIVFS NRVFNVSKPL TDPSFYPPSD PKIL RHFNI CCSTMMYLST ALGDVPSFAR LHDLYNRPIT YYFRKQVIRG NVYLSWSWSN DTSVFKRVAC NSSLSLSSHW IRLIY KIVK TTRLVGSIKD LSREVERHLH RYNRWITLED IRSRSSLLDY SCL UniProtKB: Large structural protein |
-Macromolecule #2: Phosphoprotein,Phosphoprotein
| Macromolecule | Name: Phosphoprotein,Phosphoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rabies virus (strain SAD B19) / Strain: SAD B19 Rabies virus (strain SAD B19) / Strain: SAD B19 |
| Molecular weight | Theoretical: 4.623038 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)EDMGR LHLDDGKSPN HGEIAKVGEG KYREDFQMDE GE UniProtKB: Phosphoprotein |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 8.5 |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 72.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)