+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20751 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of tetrameric sIgA complex (Class 2) | |||||||||
 Map data Map data | tetrameric sIgA complex (Class 2) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | immunoglobulin / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationpolymeric immunoglobulin receptor activity / immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor / polymeric immunoglobulin binding / dimeric IgA immunoglobulin complex / secretory dimeric IgA immunoglobulin complex / monomeric IgA immunoglobulin complex / pentameric IgM immunoglobulin complex / secretory IgA immunoglobulin complex / Fc receptor signaling pathway / IgA binding ...polymeric immunoglobulin receptor activity / immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor / polymeric immunoglobulin binding / dimeric IgA immunoglobulin complex / secretory dimeric IgA immunoglobulin complex / monomeric IgA immunoglobulin complex / pentameric IgM immunoglobulin complex / secretory IgA immunoglobulin complex / Fc receptor signaling pathway / IgA binding / IgA immunoglobulin complex / glomerular filtration / detection of chemical stimulus involved in sensory perception of bitter taste / immunoglobulin receptor binding / immunoglobulin complex, circulating / azurophil granule membrane / receptor clustering / positive regulation of respiratory burst / humoral immune response / complement activation, classical pathway / Scavenging of heme from plasma / antigen binding / Cell surface interactions at the vascular wall / B cell receptor signaling pathway / epidermal growth factor receptor signaling pathway / transmembrane signaling receptor activity / antibacterial humoral response / protein-containing complex assembly / blood microparticle / protein-macromolecule adaptor activity / adaptive immune response / receptor complex / immune response / innate immune response / Neutrophil degranulation / signal transduction / protein homodimerization activity / extracellular space / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Kumar N / Arthur CP | |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Structure of the secretory immunoglobulin A core. Authors: Nikit Kumar / Christopher P Arthur / Claudio Ciferri / Marissa L Matsumoto /  Abstract: Secretory immunoglobulin A (sIgA) represents the immune system's first line of defense against mucosal pathogens. IgAs are transported across the epithelium, as dimers and higher-order polymers, by ...Secretory immunoglobulin A (sIgA) represents the immune system's first line of defense against mucosal pathogens. IgAs are transported across the epithelium, as dimers and higher-order polymers, by the polymeric immunoglobulin receptor (pIgR). Upon reaching the luminal side, sIgAs mediate host protection and pathogen neutralization. In recent years, an increasing amount of attention has been given to IgA as a novel therapeutic antibody. However, despite extensive studies, sIgA structures have remained elusive. Here, we determine the atomic resolution structures of dimeric, tetrameric, and pentameric IgA-Fc linked by the joining chain (JC) and in complex with the secretory component of the pIgR. We suggest a mechanism in which the JC templates IgA oligomerization and imparts asymmetry for pIgR binding and transcytosis. This framework will inform the design of future IgA-based therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20751.map.gz emd_20751.map.gz | 200.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20751-v30.xml emd-20751-v30.xml emd-20751.xml emd-20751.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20751.png emd_20751.png | 129.3 KB | ||
| Filedesc metadata |  emd-20751.cif.gz emd-20751.cif.gz | 5.9 KB | ||
| Others |  emd_20751_additional.map.gz emd_20751_additional.map.gz | 199.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20751 http://ftp.pdbj.org/pub/emdb/structures/EMD-20751 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20751 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20751 | HTTPS FTP |
-Related structure data
| Related structure data |  6ue9MC  6ue7C  6ue8C  6ueaC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20751.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20751.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | tetrameric sIgA complex (Class 2) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0188 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
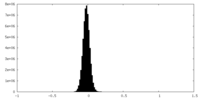
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: -150 sharpened map of tetrameric sIgA (Class 2).
| File | emd_20751_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | -150 sharpened map of tetrameric sIgA (Class 2). | ||||||||||||
| Projections & Slices |
| ||||||||||||
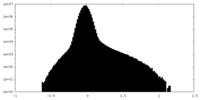
| Density Histograms |
- Sample components
Sample components
-Entire : tetrameric sIgA complex (Class 2)
| Entire | Name: tetrameric sIgA complex (Class 2) |
|---|---|
| Components |
|
-Supramolecule #1: tetrameric sIgA complex (Class 2)
| Supramolecule | Name: tetrameric sIgA complex (Class 2) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2-#3, #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 294 KDa |
-Macromolecule #1: Immunoglobulin heavy constant alpha 2
| Macromolecule | Name: Immunoglobulin heavy constant alpha 2 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.682146 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDKLV PRGSCHPRLS LHRPALEDLL LGSEANLTCT LTGLRDASGA TFTWTPSSGK SAVQGPPERD LCGCYSVSSV LPGCAQPWN HGETFTCTAA HPELKTPLTA NITKSGNTFR PEVHLLPPPS EELALNELVT LTCLARGFSP KDVLVRWLQG S QELPREKY ...String: DYKDDDDKLV PRGSCHPRLS LHRPALEDLL LGSEANLTCT LTGLRDASGA TFTWTPSSGK SAVQGPPERD LCGCYSVSSV LPGCAQPWN HGETFTCTAA HPELKTPLTA NITKSGNTFR PEVHLLPPPS EELALNELVT LTCLARGFSP KDVLVRWLQG S QELPREKY LTWASRQEPS QGTTTYAVTS ILRVAAEDWK KGETFSCMVG HEALPLAFTQ KTIDRLAGKP THINVSVVMA EA DGTCY UniProtKB: Immunoglobulin heavy constant alpha 2 |
-Macromolecule #2: Polymeric immunoglobulin receptor
| Macromolecule | Name: Polymeric immunoglobulin receptor / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 65.154094 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KSPIFGPEEV NSVEGNSVSI TCYYPPTSVN RHTRKYWCRQ GARGGCITLI SSEGYVSSKY AGRANLTNFP ENGTFVVNIA QLSQDDSGR YKCGLGINSR GLSFDVSLEV SQGPGLLNDT KVYTVDLGRT VTINCPFKTE NAQKRKSLYK QIGLYPVLVI D SSGYVNPN ...String: KSPIFGPEEV NSVEGNSVSI TCYYPPTSVN RHTRKYWCRQ GARGGCITLI SSEGYVSSKY AGRANLTNFP ENGTFVVNIA QLSQDDSGR YKCGLGINSR GLSFDVSLEV SQGPGLLNDT KVYTVDLGRT VTINCPFKTE NAQKRKSLYK QIGLYPVLVI D SSGYVNPN YTGRIRLDIQ GTGQLLFSVV INQLRLSDAG QYLCQAGDDS NSNKKNADLQ VLKPEPELVY EDLRGSVTFH CA LGPEVAN VAKFLCRQSS GENCDVVVNT LGKRAPAFEG RILLNPQDKD GSFSVVITGL RKEDAGRYLC GAHSDGQLQE GSP IQAWQL FVNEESTIPR SPTVVKGVAG GSVAVLCPYN RKESKSIKYW CLWEGAQNGR CPLLVDSEGW VKAQYEGRLS LLEE PGNGT FTVILNQLTS RDAGFYWCLT NGDTLWRTTV EIKIIEGEPN LKVPGNVTAV LGETLKVPCH FPCKFSSYEK YWCKW NNTG CQALPSQDEG PSKAFVNCDE NSRLVSLTLN LVTRADEGWY WCGVKQGHFY GETAAVYVAV EERKAAGSRD VSLAKA DAA PDEKVLDSGF REIENKAIQD PRHHHHHH UniProtKB: Polymeric immunoglobulin receptor |
-Macromolecule #3: Immunoglobulin J chain
| Macromolecule | Name: Immunoglobulin J chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.611458 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QEDERIVLVD NKCKCARITS RIIRSSEDPN EDIVERNIRI IVPLNNRENI SDPTSPLRTR FVYHLSDLCK KCDPTEVELD NQIVTATQS NICDEDSATE TCYTYDRNKC YTAVVPLVYG GETKMVETAL TPDACYPD UniProtKB: Immunoglobulin J chain |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 7 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 10.0 sec. / Average electron dose: 51.91 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cisTEM / Number images used: 144523 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: OTHER / Software - Name: cisTEM |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)