[English] 日本語
 Yorodumi
Yorodumi- EMDB-2071: Gating movement in acetylcholine receptor analysed by time-resolv... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2071 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Gating movement in acetylcholine receptor analysed by time-resolved electron cryo-microscopy | |||||||||
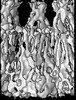
 Map data Map data | Density map of acetylcholine receptor | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | acetylcholine receptor / freeze-trapping / asymmetric gating / allosteric mechanism | |||||||||
| Function / homology |  Function and homology information Function and homology informationacetylcholine-gated channel complex / acetylcholine-gated monoatomic cation-selective channel activity / acetylcholine receptor signaling pathway / transmembrane signaling receptor activity / postsynaptic membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 6.2 Å | |||||||||
 Authors Authors | Unwin N / Fujiyoshi Y | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2012 Journal: J Mol Biol / Year: 2012Title: Gating movement of acetylcholine receptor caught by plunge-freezing. Authors: Nigel Unwin / Yoshinori Fujiyoshi /   Abstract: The nicotinic acetylcholine (ACh) receptor converts transiently to an open-channel form when activated by ACh released into the synaptic cleft. We describe here the conformational change underlying ...The nicotinic acetylcholine (ACh) receptor converts transiently to an open-channel form when activated by ACh released into the synaptic cleft. We describe here the conformational change underlying this event, determined by electron microscopy of ACh-sprayed and freeze-trapped postsynaptic membranes. ACh binding to the α subunits triggers a concerted rearrangement in the ligand-binding domain, involving an ~1-Å outward displacement of the extracellular portion of the β subunit where it interacts with the juxtaposed ends of α-helices shaping the narrow membrane-spanning pore. The β-subunit helices tilt outward to accommodate this displacement, destabilising the arrangement of pore-lining helices, which in the closed channel bend inward symmetrically to form a central hydrophobic gate. Straightening and tangential motion of the pore-lining helices effect channel opening by widening the pore asymmetrically and increasing its polarity in the region of the gate. The pore-lining helices of the α(γ) and δ subunits, by flexing between alternative bent and straight conformations, undergo the greatest movements. This coupled allosteric transition shifts the structure from a tense (closed) state toward a more relaxed (open) state. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2071.map.gz emd_2071.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2071-v30.xml emd-2071-v30.xml emd-2071.xml emd-2071.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd-2071.tif emd-2071.tif | 185.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2071 http://ftp.pdbj.org/pub/emdb/structures/EMD-2071 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2071 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2071 | HTTPS FTP |
-Validation report
| Summary document |  emd_2071_validation.pdf.gz emd_2071_validation.pdf.gz | 257.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2071_full_validation.pdf.gz emd_2071_full_validation.pdf.gz | 256.4 KB | Display | |
| Data in XML |  emd_2071_validation.xml.gz emd_2071_validation.xml.gz | 4.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2071 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2071 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2071 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2071 | HTTPS FTP |
-Related structure data
| Related structure data |  4aq5MC  2072C  4aq9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2071.map.gz / Format: CCP4 / Size: 10.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2071.map.gz / Format: CCP4 / Size: 10.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density map of acetylcholine receptor | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : nicotinic acetylcholine receptor in native postsynaptic membrane ...
| Entire | Name: nicotinic acetylcholine receptor in native postsynaptic membrane from Torpedo marmorata |
|---|---|
| Components |
|
-Supramolecule #1000: nicotinic acetylcholine receptor in native postsynaptic membrane ...
| Supramolecule | Name: nicotinic acetylcholine receptor in native postsynaptic membrane from Torpedo marmorata type: sample / ID: 1000 / Oligomeric state: 5 subunits / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 300 KDa / Theoretical: 300 KDa Method: molecular weight based on amino acid sequence data and attached sugars |
-Macromolecule #1: nicotinic acetylcholine receptor
| Macromolecule | Name: nicotinic acetylcholine receptor / type: protein_or_peptide / ID: 1 / Name.synonym: nicotinic receptor Details: Protein is embedded in postsynaptic membrane isolated from Torpedo marmorata electric organ Oligomeric state: pentamer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 300 KDa / Theoretical: 300 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Details: 100 mM sodium cacodylate, 1 mM calcium chloride |
|---|---|
| Grid | Details: 300 mesh copper grid with pre-irradiated thick holey carbon support, glow discharged in amylamine atmosphere |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 120 K / Instrument: HOMEMADE PLUNGER Details: Vitrification carried out at an ambient temperature of 8 degrees C Timed resolved state: Vitrified within 10ms of exposure to acetylcholine (applied as the grid is being plunged,using a fine, focussed spray positioned about 1cm above the ethane surface) Method: Blot until applied droplet loses contact with filter paper (indicated by loss of transparency; typically 6s) |
| Details | Tubular membrane crystals of acetylcholine receptors grow spontaneously from isolated postsynaptic membranes when incubated in low salt buffer at 17 degrees C for two weeks |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3000SFF |
|---|---|
| Temperature | Min: 10 K / Max: 20 K / Average: 10 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected based on appearance of carbon film at 250,000 times magnification |
| Details | Standard low dose imaging of specimens over holes in the carbon support film |
| Date | Nov 1, 2005 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 2.5 µm / Number real images: 111 / Average electron dose: 25 e/Å2 Details: All images recorded on film, developed in Kodak d19 developer Od range: 1 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 38500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.6 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 40000 |
| Sample stage | Specimen holder: Top-entry holder for liquid helium cooled stage (the temperature of the specimen in this holder is usually at 4K) Specimen holder model: OTHER |
- Image processing
Image processing
| Details | Alignment and distortion correction of each tube image was done using a segmental Fourier-Bessel method (Beroukhim & Unwin (1997) Ultramicroscopy, 70:57-81) with 50% overlap between successive segments |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 6.2 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: MRC, and, own, programs Details: Final maps were calculated from 111 tube images(closed class) and 123 tube images (open class) |
| CTF correction | Details: Each tube image |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: C / Chain - #3 - Chain ID: D / Chain - #4 - Chain ID: E |
|---|---|
| Software | Name: DireX |
| Details | Protocol: Maximisation of correlation between experimental densities and atomic model, using a deformable elastic network algorithm. Identical refinement procedures were applied to both density maps. The fits were validated by applying the same refinement procedures to independent density maps calculated from half-datasets |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-4aq5: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)