+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20642 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV-1 B41 SOSIP.664 in complex with rabbit antibody 13B | ||||||||||||
 Map data Map data | sharpened map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HIV-1 / rabbit antibody / SOSIP / VIRAL PROTEIN-IMMUNE SYSTEM complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / membrane Similarity search - Function | ||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  | ||||||||||||
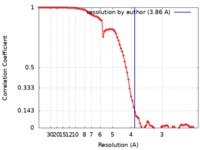
| Method | single particle reconstruction / cryo EM / Resolution: 3.86 Å | ||||||||||||
 Authors Authors | Yang YR / Ward AB | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Virol / Year: 2020 Journal: J Virol / Year: 2020Title: Autologous Antibody Responses to an HIV Envelope Glycan Hole Are Not Easily Broadened in Rabbits. Authors: Yuhe R Yang / Laura E McCoy / Marit J van Gils / Raiees Andrabi / Hannah L Turner / Meng Yuan / Christopher A Cottrell / Gabriel Ozorowski / James Voss / Matthias Pauthner / Thomas M ...Authors: Yuhe R Yang / Laura E McCoy / Marit J van Gils / Raiees Andrabi / Hannah L Turner / Meng Yuan / Christopher A Cottrell / Gabriel Ozorowski / James Voss / Matthias Pauthner / Thomas M Polveroni / Terrence Messmer / Ian A Wilson / Rogier W Sanders / Dennis R Burton / Andrew B Ward /    Abstract: Extensive studies with subtype A BG505-derived HIV envelope glycoprotein (Env) immunogens have revealed that the dominant autologous neutralizing epitope in rabbits is located in an exposed region of ...Extensive studies with subtype A BG505-derived HIV envelope glycoprotein (Env) immunogens have revealed that the dominant autologous neutralizing epitope in rabbits is located in an exposed region of the heavily glycosylated trimer that lacks potential N-linked glycosylation sites at positions 230, 241, and 289. The Env derived from B41, a subtype B virus, shares a glycan hole centered on positions 230 and 289. To test whether broader neutralization to the common glycan hole can be achieved, we immunized rabbits with B41 SOSIP (gp120-gp41 disulfide [SOS] with an isoleucine-to-proline mutation [IP] in gp41) alone, as well as B41 and BG505 coimmunization. We isolated autologous neutralizing antibodies (nAbs) and described their structure in complex with the B41 Env. Our data suggest that distinct autologous nAb lineages are induced by BG505 and B41 immunogens, even when both were administered together. In contrast to previously described BG505 glycan hole antibodies, the B41-specific nAbs accommodate the >97% conserved N241 glycan, which is present in B41. Single-particle cryo-electron microscopy studies confirmed that B41- and BG505-specific nAbs bind to overlapping glycan hole epitopes. We then used our high-resolution data to guide mutations in the BG505 glycan hole epitope in an attempt to broaden the reactivity of a B41-specific nAb, but we recovered only partial binding. Our data demonstrate that the lack of cross-reactivity in glycan hole antibodies is due to amino acid differences within the epitope, and our attempts to rationally design cross-reactive trimers resulted in only limited success. Thus, even for the immunodominant glycan hole shared between BG505 and B41, the prospect of designing prime-boost immunogens remains difficult. A glycan hole is one of the most dominant autologous neutralizing epitopes targeted on BG505 and B41 SOSIP trimer-immunized rabbits. Our high-resolution cryo-electron microscopy (cryoEM) studies of B41 in complex with a B41-specific antibody complex elucidate the molecular basis of this strain-specific glycan hole response. We conclude that even for the immunodominant glycan hole shared between BG505 and B41, the prospect of designing prime-boost immunogens remains difficult. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20642.map.gz emd_20642.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20642-v30.xml emd-20642-v30.xml emd-20642.xml emd-20642.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20642_fsc.xml emd_20642_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_20642.png emd_20642.png | 155.1 KB | ||
| Masks |  emd_20642_msk_1.map emd_20642_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20642.cif.gz emd-20642.cif.gz | 7 KB | ||
| Others |  emd_20642_half_map_1.map.gz emd_20642_half_map_1.map.gz emd_20642_half_map_2.map.gz emd_20642_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20642 http://ftp.pdbj.org/pub/emdb/structures/EMD-20642 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20642 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20642 | HTTPS FTP |
-Related structure data
| Related structure data |  6u59MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20642.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20642.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20642_msk_1.map emd_20642_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_20642_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_20642_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 B41 SOSIP.664 in complex with rabbit antibody 13B
| Entire | Name: HIV-1 B41 SOSIP.664 in complex with rabbit antibody 13B |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 B41 SOSIP.664 in complex with rabbit antibody 13B
| Supramolecule | Name: HIV-1 B41 SOSIP.664 in complex with rabbit antibody 13B type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Molecular weight | Theoretical: 570 KDa |
-Supramolecule #2: HIV-1 B41 SOSIP.664
| Supramolecule | Name: HIV-1 B41 SOSIP.664 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: B41 Human immunodeficiency virus 1 / Strain: B41 |
-Supramolecule #3: rabbit antibody 13B
| Supramolecule | Name: rabbit antibody 13B / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: SOSIP.664 gp120,SOSIP.664 gp120
| Macromolecule | Name: SOSIP.664 gp120,SOSIP.664 gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 58.872902 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAAKKW VTVYYGVPVW KEATTTLFCA SDAKAYDTEV HNVWATHACV PTDPNPQEI VLGNVTENFN MWKNNMVEQM HEDIISLWDQ SLKPCVKLTP LCVTLNCNNV NTNNTNNSTN ATISDWEKME T GEMKNCSF ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAAKKW VTVYYGVPVW KEATTTLFCA SDAKAYDTEV HNVWATHACV PTDPNPQEI VLGNVTENFN MWKNNMVEQM HEDIISLWDQ SLKPCVKLTP LCVTLNCNNV NTNNTNNSTN ATISDWEKME T GEMKNCSF NVTTSIRDKI KKEYALFYKL DVVPLENKNN INNTNITNYR LINCNTSVIT QACPKVSFEP IPIHYCAPAG FA ILKCNSK TFNGSGPCTN VSTVQCTHGI RPVVSTQLLL NGSLAEEEIV IRSENITDNA KTIIVQLNEA VEINCTRPNN NTR KSIHIG PGRAFYATGD IIGNIRQAHC NISKARWNET LGQIVAKLEE QFPNKTIIFN HSSGGDPEIV THSFNCGGEF FYCN TTPLF NSTWNNTRTD DYPTGGEQNI TLQCRIKQII NMWQGVGKAM YAPPIRGQIR CSSNITGLLL TRDGGRDQNG TETFR PGGG NMRDNWRSEL YKYKVVKIEP LGIAPTACKR RVVQRRRRRR UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: SOSIP.664 gp41
| Macromolecule | Name: SOSIP.664 gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: B41 Human immunodeficiency virus 1 / Strain: B41 |
| Molecular weight | Theoretical: 17.357824 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGLGAFILG FLGAAGSTMG AASMALTVQA RLLLSGIVQQ QNNLLRAPEA QQHMLQLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KIICCTNVPW NDSWSNKTIN EIWDNMTWMQ WEKEIDNYTQ HIYTLLEVSQ IQQEKNEQEL LELD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: rabbit antibody 13B Fragment antigen binding light chain
| Macromolecule | Name: rabbit antibody 13B Fragment antigen binding light chain type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.977202 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQTPAS VSEPVGGTVT INCQASQSRG NNYLSWYQQK PGQSPSLLIY RTSTLASGVP SRFKGSGSGT QFTLTISDLE CADAATYYC LYGYYSSRNP DFAFGGGTEV VVK |
-Macromolecule #4: rabbit antibody 13B Fragment antigen binding heavy chain
| Macromolecule | Name: rabbit antibody 13B Fragment antigen binding heavy chain type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.790192 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LEESGGGLVQ PEGSLTLTCK ASGFDFSDYH VQWVRQSPGK GLEFIGGIAY TGNIYYASWA KGRFTISKTS STTVTLQMTT LTAADTATY FCARAYGYAS APYAQYFNLW GPGTLVTVSS |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 30 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: Detergent was added to sample shortly prior to freezing. | ||||||||||||
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1721 / Average exposure time: 14.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6u59: |
 Movie
Movie Controller
Controller




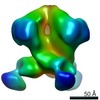
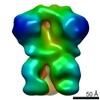

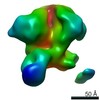


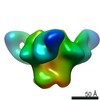






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)