+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-20629 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Best fitting antiparallel model for Volume 2 of truncated dimeric Cytohesin-3 (Grp1; amino acids 14-399) | |||||||||
 マップデータ マップデータ | Volume 2 for truncated dimeric Cytohesin-3 (Grp1; amino acids 14-399) | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Arf GEF / Phosphoinositide binding / Sec7 Domain / PH Domain / ENDOCYTOSIS | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Golgi vesicle transport / establishment of epithelial cell polarity / Intra-Golgi traffic / regulation of ARF protein signal transduction / phosphatidylinositol-3,4,5-trisphosphate binding / bicellular tight junction / ruffle / positive regulation of cell adhesion / guanyl-nucleotide exchange factor activity / adherens junction ...Golgi vesicle transport / establishment of epithelial cell polarity / Intra-Golgi traffic / regulation of ARF protein signal transduction / phosphatidylinositol-3,4,5-trisphosphate binding / bicellular tight junction / ruffle / positive regulation of cell adhesion / guanyl-nucleotide exchange factor activity / adherens junction / Golgi membrane / nucleoplasm / plasma membrane / cytosol 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / ネガティブ染色法 / 解像度: 53.0 Å | |||||||||
 データ登録者 データ登録者 | Das S / Lambright DG | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Structure / 年: 2019 ジャーナル: Structure / 年: 2019タイトル: Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes. 著者: Sanchaita Das / Andrew W Malaby / Agata Nawrotek / Wenhua Zhang / Mahel Zeghouf / Sarah Maslen / Mark Skehel / Srinivas Chakravarthy / Thomas C Irving / Osman Bilsel / Jacqueline Cherfils / David G Lambright /    要旨: Membrane dynamic processes require Arf GTPase activation by guanine nucleotide exchange factors (GEFs) with a Sec7 domain. Cytohesin family Arf GEFs function in signaling and cell migration through ...Membrane dynamic processes require Arf GTPase activation by guanine nucleotide exchange factors (GEFs) with a Sec7 domain. Cytohesin family Arf GEFs function in signaling and cell migration through Arf GTPase activation on the plasma membrane and endosomes. In this study, the structural organization of two cytohesins (Grp1 and ARNO) was investigated in solution by size exclusion-small angle X-ray scattering and negative stain-electron microscopy and on membranes by dynamic light scattering, hydrogen-deuterium exchange-mass spectrometry and guanosine diphosphate (GDP)/guanosine triphosphate (GTP) exchange assays. The results suggest that cytohesins form elongated dimers with a central coiled coil and membrane-binding pleckstrin-homology (PH) domains at opposite ends. The dimers display significant conformational heterogeneity, with a preference for compact to intermediate conformations. Phosphoinositide-dependent membrane recruitment is mediated by one PH domain at a time and alters the conformational dynamics to prime allosteric activation by Arf-GTP. A structural model for membrane targeting and allosteric activation of full-length cytohesin dimers is discussed. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_20629.map.gz emd_20629.map.gz | 162.1 KB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-20629-v30.xml emd-20629-v30.xml emd-20629.xml emd-20629.xml | 15.8 KB 15.8 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_20629.png emd_20629.png | 30.7 KB | ||
| Filedesc metadata |  emd-20629.cif.gz emd-20629.cif.gz | 6.6 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20629 http://ftp.pdbj.org/pub/emdb/structures/EMD-20629 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20629 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20629 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_20629_validation.pdf.gz emd_20629_validation.pdf.gz | 306.5 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_20629_full_validation.pdf.gz emd_20629_full_validation.pdf.gz | 306.1 KB | 表示 | |
| XML形式データ |  emd_20629_validation.xml.gz emd_20629_validation.xml.gz | 4.7 KB | 表示 | |
| CIF形式データ |  emd_20629_validation.cif.gz emd_20629_validation.cif.gz | 5.3 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20629 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20629 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20629 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20629 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_20629.map.gz / 形式: CCP4 / 大きさ: 2 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_20629.map.gz / 形式: CCP4 / 大きさ: 2 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Volume 2 for truncated dimeric Cytohesin-3 (Grp1; amino acids 14-399) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 6.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Truncated homodimer of Cytohesin-3 (Grp1, amino acids 14-399) wit...
| 全体 | 名称: Truncated homodimer of Cytohesin-3 (Grp1, amino acids 14-399) with Inositol 1,3,4,5-tetrakis phosphate (IP4) |
|---|---|
| 要素 |
|
-超分子 #1: Truncated homodimer of Cytohesin-3 (Grp1, amino acids 14-399) wit...
| 超分子 | 名称: Truncated homodimer of Cytohesin-3 (Grp1, amino acids 14-399) with Inositol 1,3,4,5-tetrakis phosphate (IP4) タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Cytohesin-3
| 分子 | 名称: Cytohesin-3 / タイプ: protein_or_peptide / ID: 1 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 46.501766 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MGHHHHHHGS PEDLSLEERE ELLDIRRRKK ELIDDIERLK YEIAEVMTEI DNLTSVEESK YTQRNAQIAM GRKKFNMDPK KGIQFLIEN DLLQSSPEDV AQFLYKGEGL NKTVIGDYLG ERDDFNIKVL QAFVELHEFA DLNLVQALRQ FLWSFRLPGE A QKIDRMME ...文字列: MGHHHHHHGS PEDLSLEERE ELLDIRRRKK ELIDDIERLK YEIAEVMTEI DNLTSVEESK YTQRNAQIAM GRKKFNMDPK KGIQFLIEN DLLQSSPEDV AQFLYKGEGL NKTVIGDYLG ERDDFNIKVL QAFVELHEFA DLNLVQALRQ FLWSFRLPGE A QKIDRMME AFASRYCLCN PGVFQSTDTC YVLSFAIIML NTSLHNHNVR DKPTAERFIT MNRGINEGGD LPEELLRNLY ES IKNEPFK IPEDDGNDLT YTFFNPDREG WLLKLGGRVK TWKRRWFILT DNCLYYFEYT TDKEPRGIIP LENLSIREVE DPR KPNCFE LYNPSHKGQV IKACKTEADG RVVEGNHVVY RISAPSPEEK EEWMKSIKAS ISRDPFYDML ATRKRRIANK K UniProtKB: Cytohesin-3 |
-分子 #2: INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE
| 分子 | 名称: INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE / タイプ: ligand / ID: 2 / コピー数: 2 / 式: 4IP |
|---|---|
| 分子量 | 理論値: 500.075 Da |
| Chemical component information | 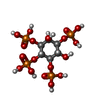 ChemComp-4IP: |
-実験情報
-構造解析
| 手法 | ネガティブ染色法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 8 構成要素:
詳細: Peak fractions after gel filtration were immediately diluted, applied to freshly glow discharged grids, and stained with uranyl formate. | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 染色 | タイプ: NEGATIVE / 材質: Uranyl Formate / 詳細: Stained with 0.75% (w/v) uranyl formate | |||||||||
| グリッド | 材質: COPPER / メッシュ: 400 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: CONTINUOUS / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 30 sec. | |||||||||
| 詳細 | The sample is a uniform homodimer with significant conformational flexibility. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI/PHILIPS CM120T |
|---|---|
| 詳細 | specimen holder: FISCHIONE INSTRUMENTS DUAL AXIS TOMOGRAPHY HOLDER |
| 撮影 | フィルム・検出器のモデル: TVIPS TEMCAM-F224 (2k x 2k) デジタル化 - サイズ - 横: 2048 pixel / デジタル化 - サイズ - 縦: 2048 pixel / 撮影したグリッド数: 1 / 実像数: 500 / 平均電子線量: 30.0 e/Å2 |
| 電子線 | 加速電圧: 120 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): -3.2 µm / 最小 デフォーカス(公称値): -1.2 µm / 倍率(公称値): 28000 |
+ 画像解析
画像解析
-原子モデル構築 1
| 初期モデル | PDB ID: Chain - Chain ID: B / Chain - Residue range: 63-399 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| 詳細 | The model with the best correlation coefficient was selected by ADP_EM from a pool of 10000 models generated by RRT_SAMPLE using rigid bodies derived from 2R09 (autoinhibited core, amino acids 63-399) and a canonical antiparallel coiled coil (amino acids 18-53) built with CCBuilder. Flexible regions with reasonable geometry were modeled with MODELLER. |
| 精密化 | 空間: REAL / プロトコル: RIGID BODY FIT / 当てはまり具合の基準: Correlation coefficient |
| 得られたモデル |  PDB-6u3g: |
 ムービー
ムービー コントローラー
コントローラー















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















