[English] 日本語
 Yorodumi
Yorodumi- EMDB-20602: Cryo-EM structure of the 48-nm repeat unit of the doublet microtu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20602 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the 48-nm repeat unit of the doublet microtubule from Tetrahymena thermophila | ||||||||||||
 Map data Map data | 48-nm repeating unit of Tetrahymena doublet | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | cilia / doublet / axoneme / microtubule inner protein / STRUCTURAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationmotile cilium / microtubule-based process / structural constituent of cytoskeleton / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / cytoskeleton / hydrolase activity / GTPase activity / GTP binding / metal ion binding / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
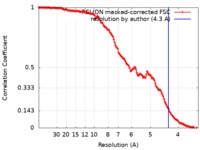
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | ||||||||||||
 Authors Authors | Ichikawa M / Khalifa AAZ / Vargas J / Basu K / Bui KH | ||||||||||||
| Funding support |  Canada, 3 items Canada, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: Tubulin lattice in cilia is in a stressed form regulated by microtubule inner proteins. Authors: Muneyoshi Ichikawa / Ahmad Abdelzaher Zaki Khalifa / Shintaroh Kubo / Daniel Dai / Kaustuv Basu / Mohammad Amin Faghfor Maghrebi / Javier Vargas / Khanh Huy Bui /   Abstract: Cilia, the hair-like protrusions that beat at high frequencies to propel a cell or move fluid around are composed of radially bundled doublet microtubules. In this study, we present a near-atomic ...Cilia, the hair-like protrusions that beat at high frequencies to propel a cell or move fluid around are composed of radially bundled doublet microtubules. In this study, we present a near-atomic resolution map of the doublet microtubule by cryoelectron microscopy. The map demonstrates that the network of microtubule inner proteins weaves into the tubulin lattice and forms an inner sheath. From mass spectrometry data and de novo modeling, we identified Rib43a proteins as the filamentous microtubule inner proteins in the protofilament ribbon region. The Rib43a-tubulin interaction leads to an elongated tubulin dimer distance every 2 dimers. In addition, the tubulin lattice structure with missing microtubule inner proteins (MIPs) by sarkosyl treatment shows significant longitudinal compaction and lateral angle change between protofilaments. These results are evidence that the MIPs directly affect and stabilize the tubulin lattice. It suggests that the doublet microtubule is an intrinsically stressed filament and that this stress could be manipulated in the regulation of ciliary waveforms. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20602.map.gz emd_20602.map.gz | 47.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20602-v30.xml emd-20602-v30.xml emd-20602.xml emd-20602.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20602_fsc.xml emd_20602_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_20602.png emd_20602.png | 131.3 KB | ||
| Filedesc metadata |  emd-20602.cif.gz emd-20602.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20602 http://ftp.pdbj.org/pub/emdb/structures/EMD-20602 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20602 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20602 | HTTPS FTP |
-Related structure data
| Related structure data |  6u0hMC  6u0tMC  6u0uMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20602.map.gz / Format: CCP4 / Size: 247.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20602.map.gz / Format: CCP4 / Size: 247.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 48-nm repeating unit of Tetrahymena doublet | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.751 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 48nm-repeat of microtubule doublet
| Entire | Name: 48nm-repeat of microtubule doublet |
|---|---|
| Components |
|
-Supramolecule #1: 48nm-repeat of microtubule doublet
| Supramolecule | Name: 48nm-repeat of microtubule doublet / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: Tubulin lattice
| Supramolecule | Name: Tubulin lattice / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Tubulin alpha chain
| Macromolecule | Name: Tubulin alpha chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.639035 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MREVISIHVG QGGIQVGNAC WELFCLEHGI QPDGQMPSDK TIGGGDDAFN TFFSETGAGK HVPRAVFLDL EPTVIDEVRT GTYRQLFHP EQLISGKEDA ANNFARGHYT IGKEIVDLCL DRIRKLADNC TGLQGFLVFN SVGGGTGSGL GSLLLERLSV D YGKKSKLG ...String: MREVISIHVG QGGIQVGNAC WELFCLEHGI QPDGQMPSDK TIGGGDDAFN TFFSETGAGK HVPRAVFLDL EPTVIDEVRT GTYRQLFHP EQLISGKEDA ANNFARGHYT IGKEIVDLCL DRIRKLADNC TGLQGFLVFN SVGGGTGSGL GSLLLERLSV D YGKKSKLG FTIYPSPQVS TAVVEPYNSI LSTHSLLEHT DVAVMLDNEA IYDICRRNLD IERPTYTNLN RLIAQVISSL TA SLRFDGA LNVDITEFQT NLVPYPRIHF MLSSYAPIIS AEKAYHEQLS VAEITNSAFE PANMMAKCDP RHGKYMACSM MYR GDVVPK DVNASIATIK TKRTIQFVDW CPTGFKVGIN YQPPTVVPGG DLAKVMRAVC MISNSTAIAE VFSRLDHKFD LMYA KRAFV HWYVGEGMEE GEFSEAREDL AALEKDYEEV GIETAEGEGE EEGY UniProtKB: Tubulin alpha chain |
-Macromolecule #2: Tubulin beta chain
| Macromolecule | Name: Tubulin beta chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.617676 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MREIVHIQGG QCGNQIGAKF WEVISDEHGI DPTGTYHGDS DLQLERINVY YNEATGGRYV PRAILMDLEP GTMDSVRAGP FGQLFRPDN FVFGQTGAGN NWAKGHYTEG AELIDSVLDV VRKEAEGCDC LQGFQITHSL GGGTGSGMGT LLISKVREEY P DRIMETFS ...String: MREIVHIQGG QCGNQIGAKF WEVISDEHGI DPTGTYHGDS DLQLERINVY YNEATGGRYV PRAILMDLEP GTMDSVRAGP FGQLFRPDN FVFGQTGAGN NWAKGHYTEG AELIDSVLDV VRKEAEGCDC LQGFQITHSL GGGTGSGMGT LLISKVREEY P DRIMETFS VVPSPKVSDT VVEPYNATLS VHQLVENADE CMVIDNEALY DICFRTLKLT TPTYGDLNHL VSAAMSGVTC CL RFPGQLN SDLRKLAVNL IPFPRLHFFM IGFAPLTSRG SQQYRALTVP ELTQQMFDAK NMMCAADPRH GRYLTASALF RGR MSTKEV DEQMLNVQNK NSSYFVEWIP NNIKSSICDI PPKGLKMAVT FVGNSTAIQE MFKRVAEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY QDATAEEEGE FEEEEGEN UniProtKB: Tubulin beta chain |
-Macromolecule #3: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force 3 for 5 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Frames/image: 1-7 / Number real images: 5983 / Average exposure time: 1.4 sec. / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.8000000000000003 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)