[English] 日本語
 Yorodumi
Yorodumi- EMDB-20450: Cryo-EM structure of the human TRPA1 ion channel in complex with ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20450 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human TRPA1 ion channel in complex with the covalent agonist BITC | |||||||||
 Map data Map data | Full map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channel / TRP channel / TRPA channel / TRPA1 channel / irritant sensing / BITC / membrane protein / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of blood circulation / positive regulation of monoatomic anion transport / cellular response to food / temperature-gated cation channel activity / regulation of neuronal action potential / cellular response to carbon dioxide / osmolarity-sensing monoatomic cation channel activity / stereocilium bundle / detection of chemical stimulus involved in sensory perception of pain / urinary bladder smooth muscle contraction ...regulation of blood circulation / positive regulation of monoatomic anion transport / cellular response to food / temperature-gated cation channel activity / regulation of neuronal action potential / cellular response to carbon dioxide / osmolarity-sensing monoatomic cation channel activity / stereocilium bundle / detection of chemical stimulus involved in sensory perception of pain / urinary bladder smooth muscle contraction / thermoception / cellular response to toxic substance / cellular response to caffeine / response to pain / TRP channels / calcium ion transmembrane import into cytosol / cellular response to cold / intracellularly gated calcium channel activity / detection of mechanical stimulus involved in sensory perception of pain / positive regulation of insulin secretion involved in cellular response to glucose stimulus / voltage-gated calcium channel activity / monoatomic ion transport / sensory perception of pain / response to cold / calcium ion transmembrane transport / calcium channel activity / cellular response to hydrogen peroxide / intracellular calcium ion homeostasis / cellular response to heat / channel activity / protein homotetramerization / cell surface receptor signaling pathway / apical plasma membrane / response to xenobiotic stimulus / axon / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||
 Authors Authors | Suo Y / Wang Z | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Neuron / Year: 2020 Journal: Neuron / Year: 2020Title: Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel. Authors: Yang Suo / Zilong Wang / Lejla Zubcevic / Allen L Hsu / Qianru He / Mario J Borgnia / Ru-Rong Ji / Seok-Yong Lee /  Abstract: Transient receptor potential channel subfamily A member 1 (TRPA1) is a Ca-permeable cation channel that serves as one of the primary sensors of environmental irritants and noxious substances. Many ...Transient receptor potential channel subfamily A member 1 (TRPA1) is a Ca-permeable cation channel that serves as one of the primary sensors of environmental irritants and noxious substances. Many TRPA1 agonists are electrophiles that are recognized by TRPA1 via covalent bond modifications of specific cysteine residues located in the cytoplasmic domains. However, a mechanistic understanding of electrophile sensing by TRPA1 has been limited due to a lack of high-resolution structural information. Here, we present the cryoelectron microscopy (cryo-EM) structures of nanodisc-reconstituted ligand-free TRPA1 and TRPA1 in complex with the covalent agonists JT010 and BITC at 2.8, 2.9, and 3.1 Å, respectively. Our structural and functional studies provide the molecular basis for electrophile recognition by the extraordinarily reactive C621 in TRPA1 and mechanistic insights into electrophile-dependent conformational changes in TRPA1. This work also provides a platform for future drug development targeting TRPA1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20450.map.gz emd_20450.map.gz | 58.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20450-v30.xml emd-20450-v30.xml emd-20450.xml emd-20450.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20450.png emd_20450.png | 210.7 KB | ||
| Filedesc metadata |  emd-20450.cif.gz emd-20450.cif.gz | 6.8 KB | ||
| Others |  emd_20450_half_map_1.map.gz emd_20450_half_map_1.map.gz emd_20450_half_map_2.map.gz emd_20450_half_map_2.map.gz | 45.8 MB 45.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20450 http://ftp.pdbj.org/pub/emdb/structures/EMD-20450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20450 | HTTPS FTP |
-Validation report
| Summary document |  emd_20450_validation.pdf.gz emd_20450_validation.pdf.gz | 901.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20450_full_validation.pdf.gz emd_20450_full_validation.pdf.gz | 900.9 KB | Display | |
| Data in XML |  emd_20450_validation.xml.gz emd_20450_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_20450_validation.cif.gz emd_20450_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20450 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20450 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20450 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20450 | HTTPS FTP |
-Related structure data
| Related structure data |  6pqpMC  6pqoC  6pqqC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20450.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20450.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.066 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: half map 1
| File | emd_20450_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
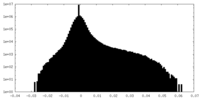
| Density Histograms |
-Half map: half map 2
| File | emd_20450_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
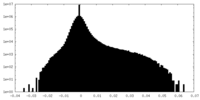
| Density Histograms |
- Sample components
Sample components
-Entire : Transient receptor potential cation channel subfamily A member 1
| Entire | Name: Transient receptor potential cation channel subfamily A member 1 |
|---|---|
| Components |
|
-Supramolecule #1: Transient receptor potential cation channel subfamily A member 1
| Supramolecule | Name: Transient receptor potential cation channel subfamily A member 1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transient receptor potential cation channel subfamily A member 1
| Macromolecule | Name: Transient receptor potential cation channel subfamily A member 1 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 131.495547 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAKRSLRKMW RPGEKKEPQG VVYEDVPDDT EDFKESLKVV FEGSAYGLQN FNKQKKLKRC DDMDTFFLHY AAAEGQIELM EKITRDSSL EVLHEMDDYG NTPLHCAVEK NQIESVKFLL SRGANPNLRN FNMMAPLHIA VQGMNNEVMK VLLEHRTIDV N LEGENGNT ...String: MAKRSLRKMW RPGEKKEPQG VVYEDVPDDT EDFKESLKVV FEGSAYGLQN FNKQKKLKRC DDMDTFFLHY AAAEGQIELM EKITRDSSL EVLHEMDDYG NTPLHCAVEK NQIESVKFLL SRGANPNLRN FNMMAPLHIA VQGMNNEVMK VLLEHRTIDV N LEGENGNT AVIIACTTNN SEALQILLKK GAKPCKSNKW GCFPIHQAAF SGSKECMEII LRFGEEHGYS RQLHINFMNN GK ATPLHLA VQNGDLEMIK MCLDNGAQID PVEKGRCTAI HFAATQGATE IVKLMISSYS GSVDIVNTTD GCHETMLHRA SLF DHHELA DYLISVGADI NKIDSEGRSP LILATASASW NIVNLLLSKG AQVDIKDNFG RNFLHLTVQQ PYGLKNLRPE FMQM QQIKE LVMDEDNDGC TPLHYACRQG GPGSVNNLLG FNVSIHSKSK DKKSPLHFAA SYGRINTCQR LLQDISDTRL LNEGD LHGM TPLHLAAKNG HDKVVQLLLK KGALFLSDHN GWTALHHASM GGYTQTMKVI LDTNLKCTDR LDEDGNTALH FAAREG HAK AVALLLSHNA DIVLNKQQAS FLHLALHNKR KEVVLTIIRS KRWDECLKIF SHNSPGNKCP ITEMIEYLPE CMKVLLD FC MLHSTEDKSC RDYYIEYNFK YLQCPLEFTK KTPTQDVIYE PLTALNAMVQ NNRIELLNHP VCKEYLLMKW LAYGFRAH M MNLGSYCLGL IPMTILVVNI KPGMAFNSTG IINETSDHSE ILDTTNSYLI KTCMILVFLS SIFGYCKEAG QIFQQKRNY FMDISNVLEW IIYTTGIIFV LPLFVEIPAH LQWQCGAIAV YFYWMNFLLY LQRFENCGIF IVMLEVILKT LLRSTVVFIF LLLAFGLSF YILLNLQDPF SSPLLSIIQT FSMMLGDINY RESFLEPYLR NELAHPVLSF AQLVSFTIFV PIVLMNLLIG L AVGDIAEV QKHASLKRIA MQVELHTSLE KKLPLWFLRK VDQKSTIVYP NKPRSGGMLF HIFCFLFCTG EIRQEIPNAD KS LEMEILK QKYRLKDLTF LLEKQHELIK LIIQKMEIIS ETEDDDSHCS FQDRFKKEQM EQRNSRWNTV LRAVKAKTHH LEP SNSLEV LFQGPAADYK DDDDKAHHHH HHHHHH UniProtKB: Transient receptor potential cation channel subfamily A member 1 |
-Macromolecule #3: N-benzylthioformamide
| Macromolecule | Name: N-benzylthioformamide / type: ligand / ID: 3 / Number of copies: 4 / Formula: 9BE |
|---|---|
| Molecular weight | Theoretical: 151.229 Da |
| Chemical component information | 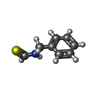 ChemComp-9BE: |
-Macromolecule #4: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyl...
| Macromolecule | Name: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate type: ligand / ID: 4 / Number of copies: 20 / Formula: 6OU |
|---|---|
| Molecular weight | Theoretical: 717.996 Da |
| Chemical component information |  ChemComp-6OU: |
-Macromolecule #5: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
| Macromolecule | Name: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine / type: ligand / ID: 5 / Number of copies: 4 / Formula: LBN |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-LBN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 26 sec. / Details: 15 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 4418 / Average exposure time: 4.6 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.25 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)